Search Count: 77,249
 |
Organism: Mycobacterium tuberculosis h37rv, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN |
 |
Organism: Mycobacterium tuberculosis h37rv, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Mycobacterium tuberculosis h37rv, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN |
 |
Organism: Mycobacterium tuberculosis h37rv, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN |
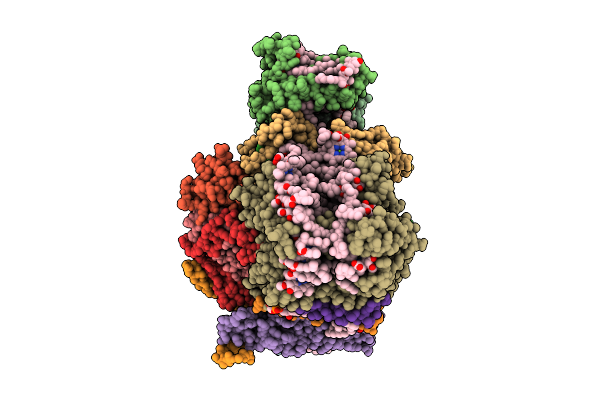 |
Structure Of The Wild-Type Psi-5Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, LMG, A1L1F, PQN, LHG, BCR, SF4 |
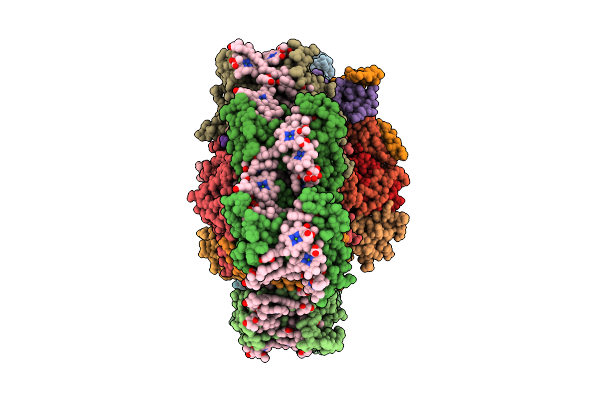 |
Structure Of The Canthaxanthin Mutant Psi-9Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, A1L1F, 45D, LHG, DGD, LMG, PQN, BCR, SF4 |
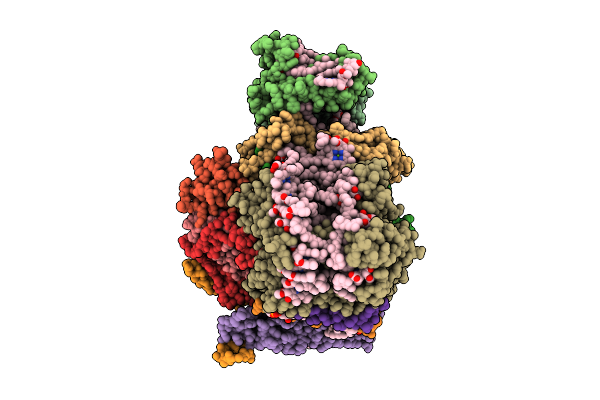 |
Structure Of The Canthaxanthin Mutant Psi-5Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, LMG, A1L1F, PQN, LHG, BCR, SF4 |
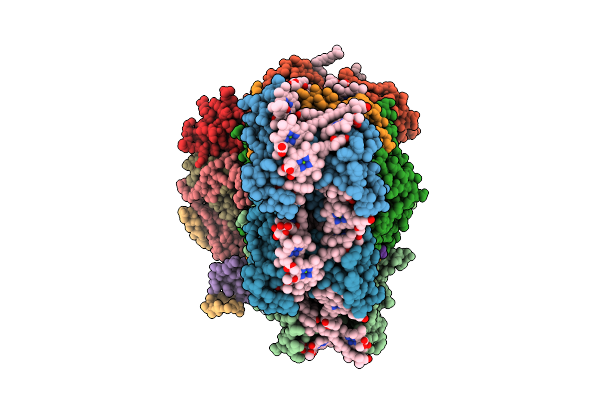 |
Structure Of The Canthaxanthin Mutant Psi-4Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: A1L1G, A1L1F, XAT, 45D, LHG, CLA, DGD, SQD, PQN, BCR, SF4, LMG |
 |
Structure Of The Canthaxanthin Mutant Psi-3Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, A1L1F, PQN, LHG, BCR, SF4, LMG |
 |
Structure Of The Astaxanthin Mutant Psi-9Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, A1L1F, LHG, DGD, LMG, PQN, BCR, SF4 |
 |
Structure Of The Astaxanthin Mutant Psi-7Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, A1L1F, LHG, DGD, PQN, BCR, SF4, LMG |
 |
Structure Of The Astaxanthin Mutant Psi-5Vcpi Supercomplex In Nannochloropsis Oceanica
Organism: Nannochloropsis oceanica strain imet1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: PHOTOSYNTHESIS Ligands: XAT, A1L1G, CLA, SQD, DGD, LMG, A1L1F, PQN, LHG, BCR, SF4 |
 |
Xfel Structure Of Hnqo1 Mixed With Nadh In An Extended Orientation At 0.3 S
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: NAD, FAD, ACT |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2025-12-10 Classification: DNA Ligands: LDP |
 |
Organism: Parastagonospora nodorum sn15
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: FAD, GOL |
 |
Solution Structure Of An Intramolecular Anti-Parallel G-Quadruplex With A 5'-End Overhang.
|
 |
Organism: Mycobacterium tuberculosis, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: DNA BINDING PROTEIN |
 |
Organism: Rouxiella badensis
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE |
 |
Organism: Mesorhizobium metallidurans
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: P6G, PEG, EPE, NA, TRS |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ELECTRON TRANSPORT Ligands: FE2, FES, HCI |

