Search Count: 52
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2024-12-18 Classification: OXIDOREDUCTASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2024-12-18 Classification: OXIDOREDUCTASE Ligands: NAP, MG |
 |
Crystal Structure Of Pyridoxal Reductase (Pdxi)In Complex With Nadph And Pyridoxal
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2024-12-18 Classification: OXIDOREDUCTASE Ligands: NAP, UEG, MG |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-03-20 Classification: LYASE |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2024-03-20 Classification: LYASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-02-07 Classification: OXIDOREDUCTASE Ligands: BME, FMN, PLP, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2024-02-07 Classification: OXIDOREDUCTASE Ligands: PO4, FMN |
 |
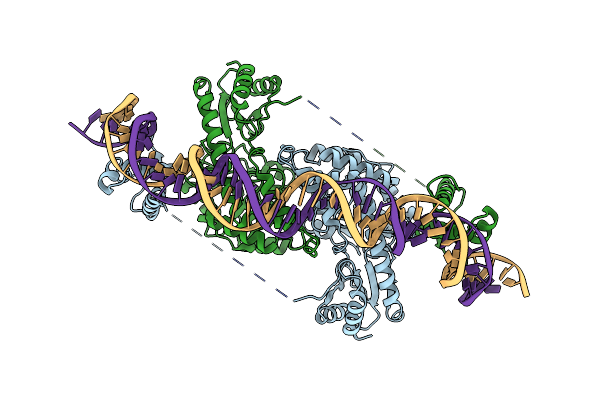
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Half-Closed Conformation
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
 |
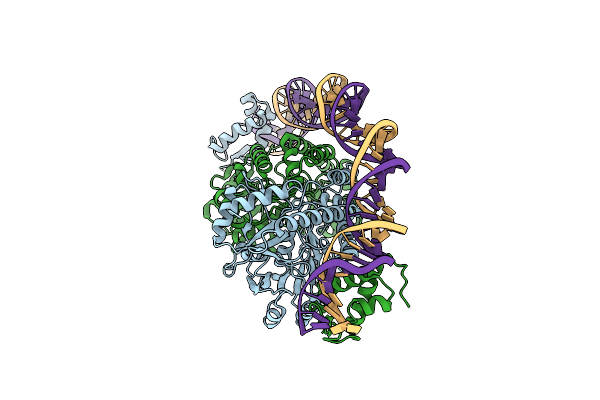
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Closed Conformation, C2 Symmetry.
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
 |
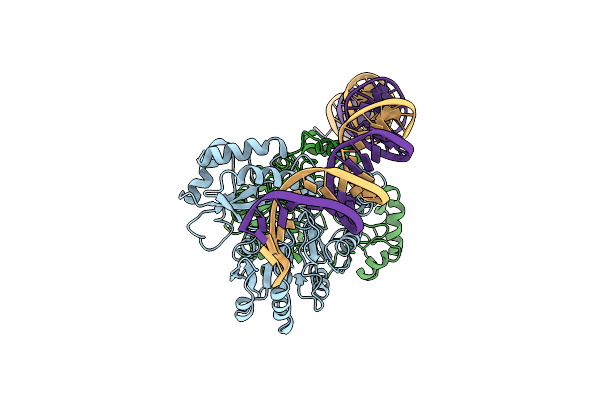
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Closed Conformation, C1 Symmetry
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
 |
Cryo-Em Structure Of Holo-Pdxr From Bacillus Clausii Bound To Its Target Dna In The Open Conformation
Organism: Alkalihalobacillus clausii
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: DNA BINDING PROTEIN |
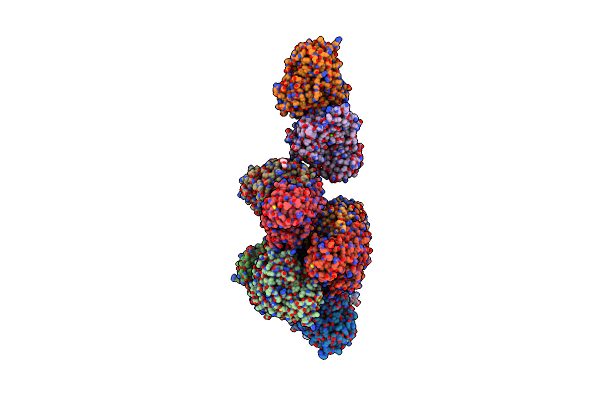 |
Organism: Alkalihalobacillus clausii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-09-28 Classification: DNA BINDING PROTEIN Ligands: CA, CL, PGE, EDO, PEG, P6G |
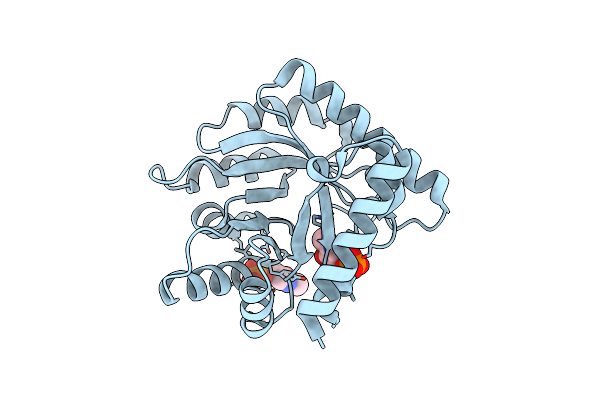 |
The Crystal Structure Of The K36A/K38A/K233A/K234A Quadruple Mutant Of E. Coli Yggs In Complex With Plp
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-03-30 Classification: PROTEIN TRANSPORT Ligands: PLP |
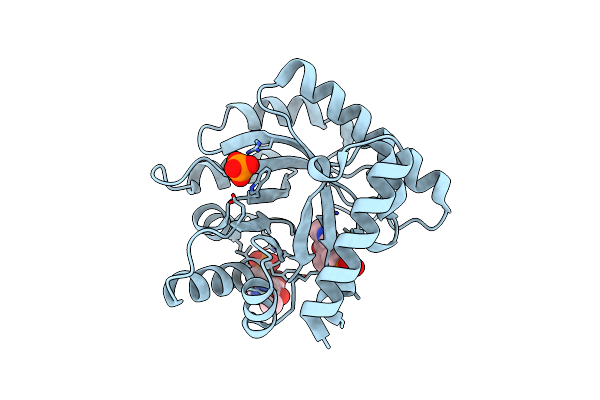 |
Crystal Structure Of The Wild Type Escherichia Coli Pyridoxal 5'-Phosphate Homeostasis Protein (Yggs)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-03-23 Classification: PROTEIN TRANSPORT Ligands: PLP, PO4 |
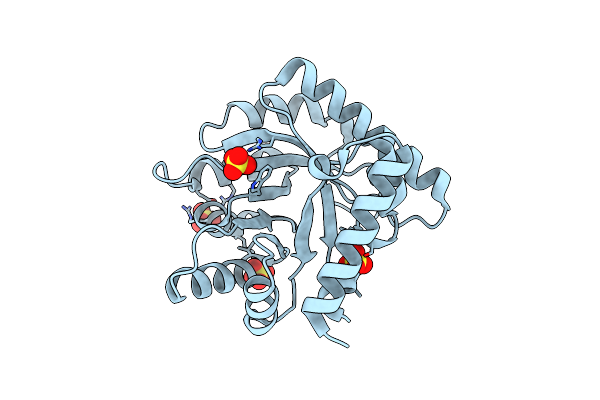 |
Crystal Structure Of Escherichia Coli Apo Pyridoxal 5'-Phosphate Homeostasis Protein (Yggs)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-03-23 Classification: PROTEIN TRANSPORT Ligands: SO4 |
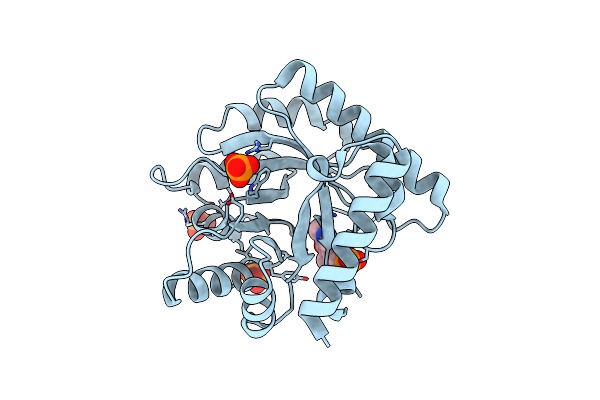 |
The Crystal Structure Of The K36A Mutant Of E. Coli Yggs In Complex With Plp
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-03-23 Classification: PROTEIN TRANSPORT Ligands: PLP, PO4 |
 |
The Crystal Structure Of The K137A Mutant Of E. Coli Yggs In Complex With Plp
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-03-23 Classification: PROTEIN TRANSPORT Ligands: PLP, SO4 |
 |
The Crystal Structure Of The K36A/K38A Double Mutant Of E. Coli Yggs In Complex With Plp
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2022-03-23 Classification: PROTEIN TRANSPORT Ligands: PO4 |
 |
The Crystal Structure Of The K38A/K137A/K233A/K234A Quadruple Mutant Of E. Coli Yggs In Complex With Plp
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-03-23 Classification: PROTEIN TRANSPORT Ligands: PLP, BU1 |
 |
The Crystal Structure Of The K36A/K137A Double Mutant Of E. Coli Yggs In Complex With Plp
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-03-23 Classification: PROTEIN TRANSPORT Ligands: PLP, SO4 |

