Search Count: 83
 |
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-07-10 Classification: UNKNOWN FUNCTION Ligands: PEG, EDO, NI |
 |
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2024-06-26 Classification: UNKNOWN FUNCTION Ligands: EDO, GOL |
 |
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2024-06-26 Classification: UNKNOWN FUNCTION |
 |
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-06-26 Classification: UNKNOWN FUNCTION |
 |
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-06-26 Classification: UNKNOWN FUNCTION Ligands: EDO, NA |
 |
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2024-06-26 Classification: UNKNOWN FUNCTION |
 |
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-06-26 Classification: UNKNOWN FUNCTION Ligands: EDO, K |
 |
Organism: Capra hircus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-06-26 Classification: UNKNOWN FUNCTION Ligands: NI, EDO, PEG |
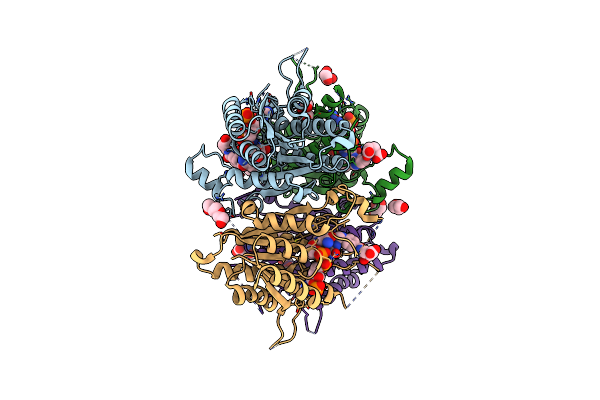 |
The Crystal Structure Of Leishmania Major Pteridine Reductase 1 In Complex With Substrate Folic Acid
Organism: Leishmania major
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2022-04-06 Classification: OXIDOREDUCTASE Ligands: FOL, NDP, PGE, GOL, PEG |
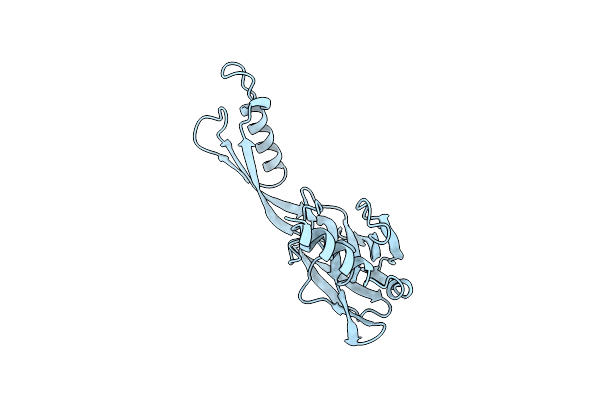 |
Organism: Trypanosoma cruzi
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2022-02-02 Classification: IMMUNE SYSTEM |
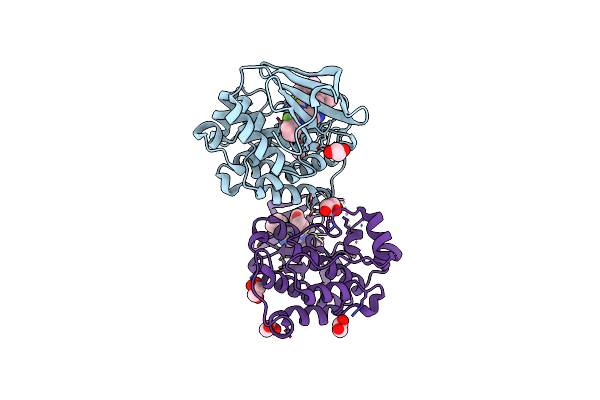 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2021-10-06 Classification: ONCOPROTEIN Ligands: RCK, EDO, DMS, GOL |
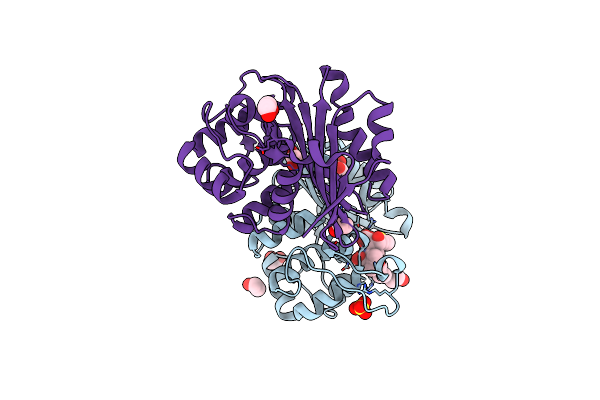 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-08-11 Classification: HYDROLASE Ligands: EDO, 8YL, SO4, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2021-07-21 Classification: HYDROLASE Ligands: QP2, EDO, SO4, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2021-07-21 Classification: HYDROLASE Ligands: 2RG, EDO |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-07-21 Classification: HYDROLASE Ligands: EDO, DWZ |
 |
Crystal Structure Of Class D Beta-Lactamase Oxa-48 In Complex With Ertapenem
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2021-07-21 Classification: HYDROLASE Ligands: 2RG, EDO, CL, PGE |
 |
Crystal Structure Of Class D Beta-Lactamase Oxa-48 In Complex With Meropenem
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2021-07-21 Classification: HYDROLASE Ligands: DWZ, EDO, CL |
 |
Organism: Ogataea glucozyma
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2021-02-10 Classification: OXIDOREDUCTASE Ligands: NAP, ACT, EPE, PGE, PEG, EDO |
 |
Organism: Alicyclobacillus herbarius
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2021-02-03 Classification: HYDROLASE Ligands: EDO, NI, PGE, PEG |
 |
Organism: Schistosoma mansoni
Method: X-RAY DIFFRACTION Resolution:3.22 Å Release Date: 2020-09-30 Classification: ALLERGEN |

