Search Count: 20
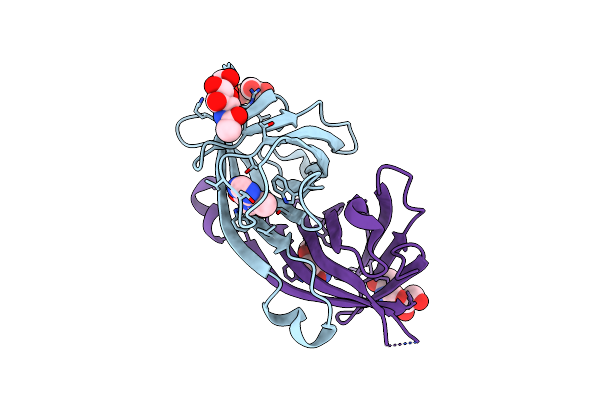 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-12-27 Classification: PROTEIN BINDING Ligands: GOL, NAG, TEP, SO4 |
 |
Crystal Structure Of Recjcdc45 From Methanothermobacter Thermoautotroficus In Complex With Dctp
Organism: Methanothermobacter thermautotrophicus (strain atcc 29096 / dsm 1053 / jcm 10044 / nbrc 100330 / delta h)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-08-03 Classification: DNA BINDING PROTEIN Ligands: MN, CTP, NO3, PEG |
 |
Crystal Structure Of Recj-Cdc45 From Methanothermobacter Thermoautotrophicus In The Closed State.
Organism: Methanothermobacter thermautotrophicus (strain atcc 29096 / dsm 1053 / jcm 10044 / nbrc 100330 / delta h)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-07-27 Classification: DNA BINDING PROTEIN Ligands: MN, PO4, PEG |
 |
Crystal Structure Of Recjcdc45 From Methanothermobacter Thermoautotroficus In Complex With Ssdna
Organism: Methanothermobacter thermautotrophicus (strain atcc 29096 / dsm 1053 / jcm 10044 / nbrc 100330 / delta h), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-07-27 Classification: DNA BINDING PROTEIN Ligands: MN |
 |
Crystal Structure Of Recj-Cdc45 From Methanothermobacter Thermoautotrophicus
Organism: Methanothermobacter thermautotrophicus (strain atcc 29096 / dsm 1053 / jcm 10044 / nbrc 100330 / delta h)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-07-20 Classification: DNA BINDING PROTEIN Ligands: MN |
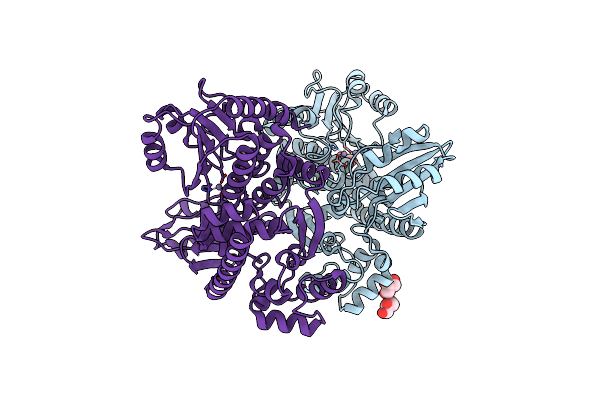 |
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-01-27 Classification: DNA BINDING PROTEIN Ligands: MN, GOL |
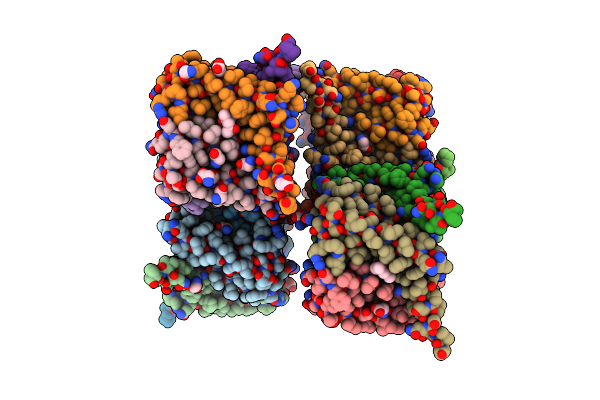 |
Crystal Structure Of Cng Mimicking Nak-Eapp Mutant (T67A) Cocrystallized With K+
Organism: Bacillus cereus (strain atcc 14579 / dsm 31 / jcm 2152 / nbrc 15305 / ncimb 9373 / nrrl b-3711), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2019-03-06 Classification: TRANSPORT PROTEIN Ligands: GLY, K, MPD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-08-22 Classification: REPLICATION |
 |
Modulation Of Pcna Sliding Surface By P15Paf Suggests A Suppressive Mechanism For Cisplatin-Induced Dna Lesion Bypass By Pol Eta Holoenzyme
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2018-08-08 Classification: DNA BINDING PROTEIN |
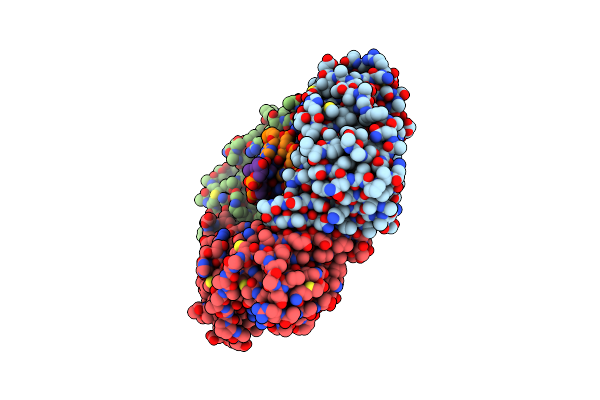 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2018-05-30 Classification: DNA BINDING PROTEIN |
 |
Organism: Bacillus cereus atcc 14579
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2015-07-01 Classification: TRANSPORT PROTEIN Ligands: MPD, GLY |
 |
Organism: Bacillus cereus atcc 14579
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-07-01 Classification: TRANSPORT PROTEIN Ligands: CS, MPD, GLY |
 |
Crystal Structure Of Cng Mimicking Nak-Etpp Mutant Cocrystallized With Dimethylammonium
Organism: Bacillus cereus atcc 14579
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-07-01 Classification: TRANSPORT PROTEIN Ligands: GLY, MPD, DMN |
 |
Organism: Bacillus cereus atcc 14579
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-07-01 Classification: TRANSPORT PROTEIN Ligands: RB, MPD, GLY |
 |
Organism: Bacillus cereus atcc 14579
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2015-07-01 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of Cng Mimicking Nak-Etpp Mutant Cocrystallized With Methylammonium
Organism: Bacillus cereus atcc 14579
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-07-01 Classification: TRANSPORT PROTEIN Ligands: 3P8, GLY, MPD |
 |
The Experimental X-Ray Structure Of The New Diheme Cytochrome Type C From Shewanella Baltica Os155 Sb-Dhc
Organism: Shewanella baltica
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2012-10-31 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Merohedral Twinning In Protein Crystals Revealed A New Synthetic Three Helix Bundle Motif
Organism: Synthetic
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2012-09-26 Classification: DE NOVO PROTEIN Ligands: SO4 |
 |
Electron Transfer Complexes: Experimental Mapping Of The Redox-Dependent Cytochrome C Electrostatic Surface
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2012-01-25 Classification: ELECTRON TRANSPORT Ligands: HEC, NO3 |
 |
Electron Transfer Complexes:Experimental Mapping Of The Redox-Dependent Cytochrome C Electrostatic Surface
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-01-25 Classification: ELECTRON TRANSPORT Ligands: HEC, NO3 |

