Search Count: 25
 |
Crystal Structure Of B1E11K Malarial Antibody In Complex With Resa Repeat Peptide
Organism: Homo sapiens, Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2024-10-30 Classification: IMMUNE SYSTEM Ligands: SO4, EDO |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2023-12-20 Classification: HYDROLASE Ligands: NAG, GOL, PEG, PG4, 1PE |
 |
Structure Of Prolyl Endoprotease From Aspergillus Niger Cbs 109712 In Space Group C222(1)
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-12-20 Classification: HYDROLASE Ligands: PG4 |
 |
Crystal Structure Of Malaria Transmission-Blocking Antigen Pfs48/45-6C Variant In Complex With Human Antibodies Rupa-44 And Rupa-29
Organism: Homo sapiens, Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2023-02-15 Classification: IMMUNE SYSTEM Ligands: NAG |
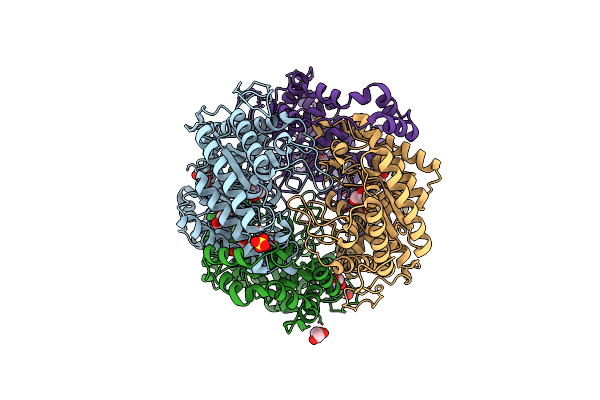 |
Crystal Structure Of Xylose Isomerase From Piromyces E2 In Complex With One Mg2+ Ions And Glycerol
Organism: Piromyces sp. e2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-11-01 Classification: ISOMERASE Ligands: MG, SO4, GOL |
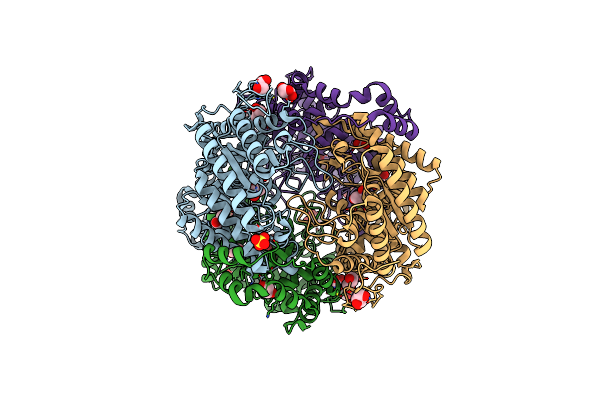 |
Organism: Piromyces sp. e2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-11-01 Classification: ISOMERASE Ligands: CA, FE2, MG, SO4, GOL |
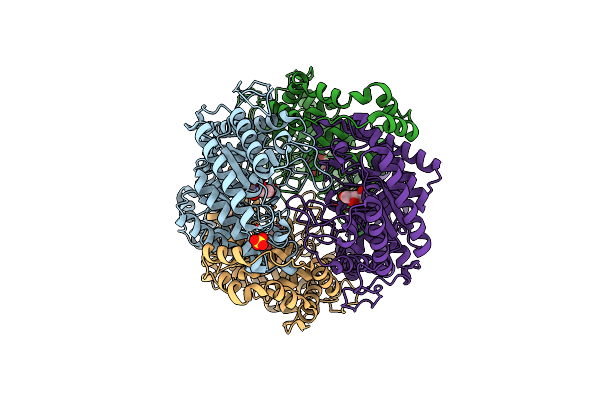 |
Crystal Structure Of Xylose Isomerase From Piromyces E2 Complexed With One Mg2+ Ion And Xylitol
Organism: Piromyces sp. e2
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2017-11-01 Classification: ISOMERASE Ligands: MG, XYL, SO4 |
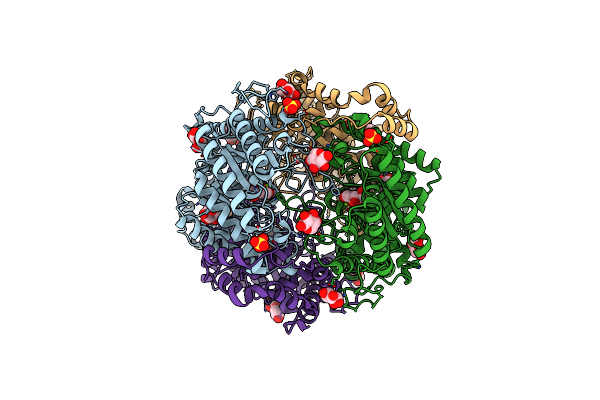 |
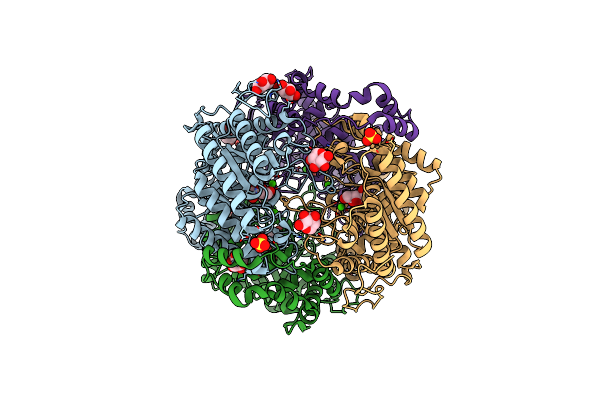
Crystal Structure Of Xylose Isomerase From Piromyces E2 In Complex With Two Mg2+ Ions And Xylose
Organism: Piromyces sp. e2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-11-01 Classification: ISOMERASE Ligands: MG, XLS, XYP, XYS, SO4 |
 |
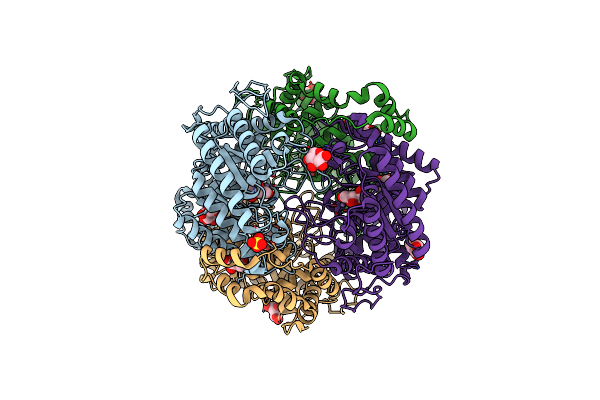
Crystal Structure Of Xylose Isomerase From Piromyces E2 In Complex With Two Ca2+ Ions And Xylose
Organism: Piromyces sp. e2
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2017-11-01 Classification: ISOMERASE Ligands: CA, XLS, XYP, SO4 |
 |
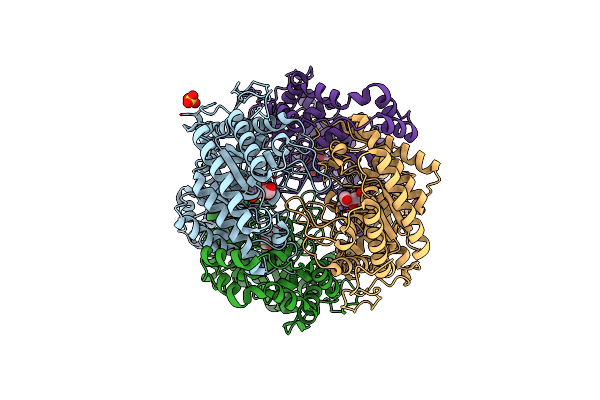
Crystal Structure Of Xylose Isomerase From Piromyces E2 In Complex With Two Mn2+ Ions And Xylose
Organism: Piromyces sp. e2
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2017-11-01 Classification: ISOMERASE Ligands: MN, XLS, XYP, SO4 |
 |
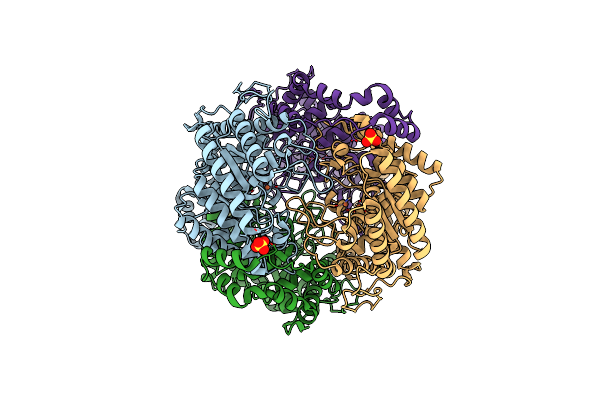
Crystal Structure Of Xylose Isomerase From Piromyces Sp. E2 In Complex With Two Mn2+ Ions And Sorbitol
Organism: Piromyces sp. e2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-11-01 Classification: ISOMERASE Ligands: MN, SOR, SO4 |
 |
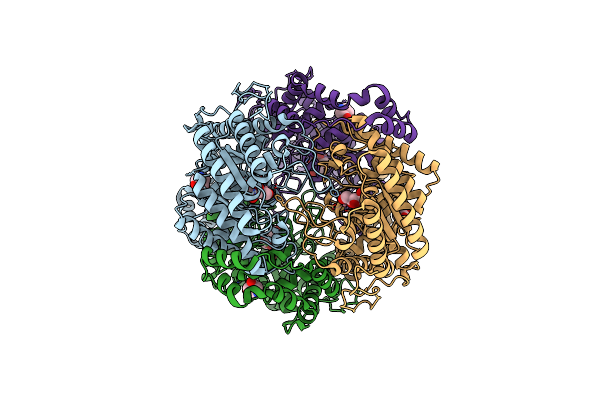
Crystal Structure Of Xylose Isomerase From Piromyces E2 In Complex With Two Fe2+ Ions
Organism: Piromyces sp. e2
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-11-01 Classification: ISOMERASE Ligands: FE2, SO4 |
 |
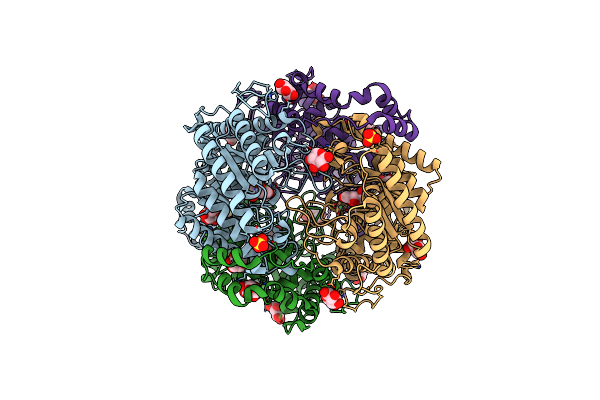
Crystal Structure Of Xylose Isomerase From Piromyces E2 In Complex With Two Co2+ Ions And Xylulose
Organism: Piromyces sp. e2
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2017-11-01 Classification: ISOMERASE Ligands: CO, XUL, HYP |
 |
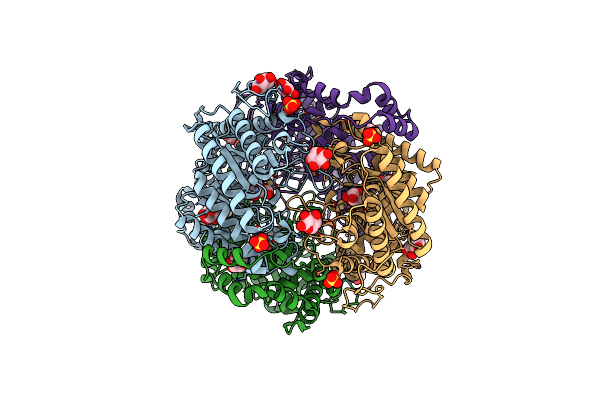
Crystal Structure Of Xylose Isomerase From Piromyces E2 In Complex With 2 Ni2+ Ions And Xylose
Organism: Piromyces sp. e2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-11-01 Classification: ISOMERASE Ligands: NI, XLS, XYP, XYS, SO4 |
 |
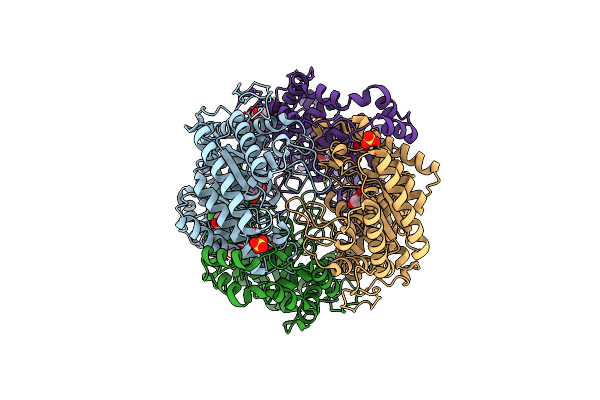
Crystal Structure Of Xylose Isomerase From Piromyces E2 In Complex With Two Cd2+ Ions And Xylose
Organism: Piromyces sp. e2
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2017-11-01 Classification: ISOMERASE Ligands: CD, XLS, XYP, XYS, SO4 |
 |
Organism: Piromyces sp. (strain e2)
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2017-11-01 Classification: ISOMERASE Ligands: GOL, SO4, ACY |
 |
The X-Ray Structure Of The Cis-3-Chloroacrylic Acid Dehalogenase Cis-Caad Inactivated With (R)-Oxirane-2-Carboxylate
Organism: Coryneform bacterium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-11-21 Classification: HYDROLASE Ligands: LAC |
 |
The X-Ray Structure Of Cis-3-Chloroacrylic Acid Dehalogenase (Cis-Caad) With A Sulfate Ion Bound In The Active Site
Organism: Coryneform bacterium
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2006-11-21 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Arthrobacter sp. ad2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-04-25 Classification: LYASE |
 |
Structure Of Haloalcohol Dehalogenase Hhec Of Agrobacterium Radiobacter Ad1 In Complex With (R)-Para-Nitro Styrene Oxide, With A Water Molecule In The Halide-Binding Site
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2005-10-04 Classification: LYASE Ligands: RNO |

