Search Count: 146
All
Selected
 |
Carbohydrate-Binding Module 32 Of Lnbb From Bifidobacterium Bifidum, Ligand Free Form, Multiple Small-Wedge Data Set
Organism: Bifidobacterium bifidum jcm 1254
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-11-19 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Carbohydrate-Binding Module 32 Of Lnbb From Bifidobacterium Bifidum, Lnb Complex
Organism: Bifidobacterium bifidum jcm 1254
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-08-13 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Organism: Ruminococcus bromii l2-63
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2023-06-14 Classification: SUGAR BINDING PROTEIN Ligands: EDO, SCN, CA |
 |
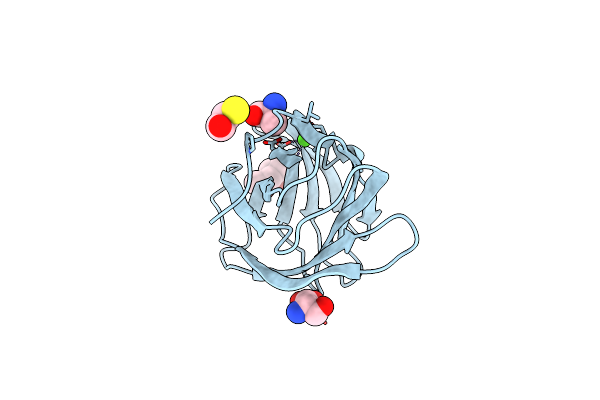
Organism: Ruminococcus bromii l2-63
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2023-06-14 Classification: SUGAR BINDING PROTEIN Ligands: EDO, SCN, CA |
 |
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-02-08 Classification: SUGAR BINDING PROTEIN Ligands: MPD, SER, BME, ALA, CA, CL |
 |
Crystal Structure Of The C-Terminal Domain Of Human N-Acetylglucosaminyltransferase Iva
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2022-09-28 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of The C-Terminal Domain Of Bombyx Mori N-Acetylglucosaminyltransferase Iv
Organism: Bombyx mori
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2022-09-28 Classification: SUGAR BINDING PROTEIN Ligands: MES, EDO |
 |
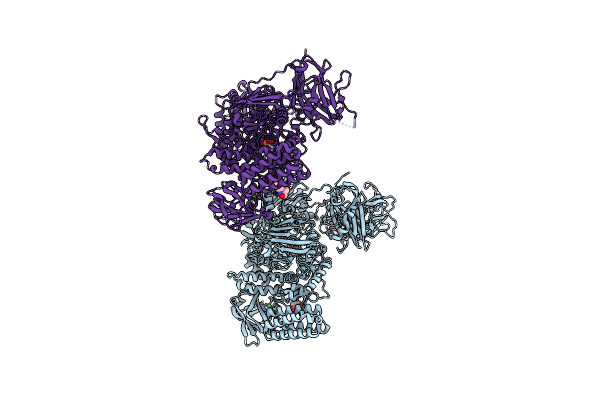
Crystal Structure Of The C-Terminal Domain Of Bombyx Mori N-Acetylglucosaminyltransferase Iv In Complex With N-Acetylglucosamine
Organism: Bombyx mori
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2022-09-28 Classification: SUGAR BINDING PROTEIN Ligands: NAG, EDO |
 |
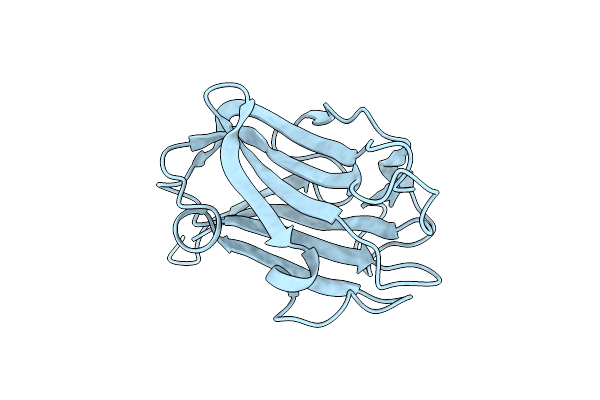
Multi-Domain Gh92 Alpha-1,2-Mannosidase From Neobacillus Novalis: Mannoimidazole Complex
Organism: Neobacillus novalis
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2022-09-21 Classification: HYDROLASE Ligands: CA, MVL, B3P |
 |
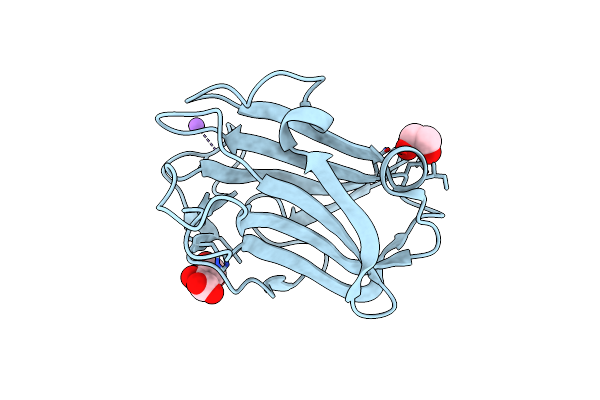
The Crystal Structure Of Family 8 Carbohydrate-Binding Module From Dictyostelium Discoideum Complexed With Iodine Atoms
Organism: Dictyostelium discoideum
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2022-04-20 Classification: SUGAR BINDING PROTEIN Ligands: IOD |
 |
The Crystal Structure Of Family 8 Carbohydrate-Binding Module From Dictyostelium Discoideum
Organism: Dictyostelium discoideum
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2022-04-20 Classification: SUGAR BINDING PROTEIN Ligands: EDO, NA, LMR |
 |
Structure Of Cbm Bt3015C From Bacteroides Thetaiotaomicron In Complex With Galactose
Organism: Bacteroides thetaiotaomicron (strain atcc 29148 / dsm 2079 / nctc 10582 / e50 / vpi-5482)
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2022-03-02 Classification: SUGAR BINDING PROTEIN Ligands: GAL, CA |
 |
Structure Of Cbm Bt3015C From Bacteroides Thetaiotaomicron In Complex With O-Galnac Core 1-Thr
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2022-03-02 Classification: SUGAR BINDING PROTEIN Ligands: THR, CA |
 |
Structure Of Cbm Bt3015C From Bacteroides Thetaiotaomicron In Complex With O-Galnac Core 2-Thr
Organism: Bacteroides thetaiotaomicron (strain atcc 29148 / dsm 2079 / nctc 10582 / e50 / vpi-5482)
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2022-03-02 Classification: SUGAR BINDING PROTEIN Ligands: EDO, THR, CA |
 |
Organism: Bacteroides thetaiotaomicron (strain atcc 29148 / dsm 2079 / nctc 10582 / e50 / vpi-5482)
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2022-03-02 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Nmr Solution Structure Of The Carbohydrate-Binding Module Family 5 (Cbm5) From Cellvibrio Japonicus Cjlpmo10A
Organism: Cellvibrio japonicus (strain ueda107)
Method: SOLUTION NMR Release Date: 2021-05-05 Classification: SUGAR BINDING PROTEIN |
 |
Nmr Solution Structure Of The Carbohydrate-Binding Module Family 73 (Cbm73) From Cellvibrio Japonicus Cjlpmo10A
Organism: Cellvibrio japonicus ueda107
Method: SOLUTION NMR Release Date: 2021-05-05 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-12-23 Classification: PEPTIDE BINDING PROTEIN Ligands: PG4, GOL, SO4 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-12-23 Classification: PEPTIDE BINDING PROTEIN Ligands: GOL |
 |
Crystal Structure Of The Cbm3 From Bacillus Subtilis At 1.06 Angstrom Resolution
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: PEG |

