Search Count: 20
 |
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2016-06-08 Classification: TRANSFERASE Ligands: FAD, PGE, DUR |
 |
Flavin-Dependent Thymidylate Synthase In Complex With Fad And 2'-Deoxyuridine-5'-Monosulfate
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2016-06-08 Classification: TRANSFERASE Ligands: FAD, DUS, RBF |
 |
Flavin-Dependent Thymidylate Synthase R90A Variant In Complex With Fad And Deoxyuridine Monophosphate
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-06-08 Classification: TRANSFERASE Ligands: FAD, UMP |
 |
Flavin-Dependent Thymidylate Synthase R174A Variant In Complex With Fad And Dump
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-06-08 Classification: TRANSFERASE Ligands: FAD, UMP |
 |
Solution Structure Of A De Novo Designed Peptide That Sequesters Toxic Heavy Metals
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2015-04-01 Classification: DE NOVO PROTEIN |
 |
Nmr-Rdc / Xray Structure Of E. Coli Hsp70 (Dnak) Chaperone (1-605) Complexed With Adp And Substrate
|
 |
Nmr Structure Of The Substrate Binding Domain Of Dnak Bound To The Peptide Nrllltg
|
 |
Best 20 Nmr Conformers Of D130I Mutant T3-I2, A 32 Residue Peptide From The Alpha 2A Adrenergic Receptor
|
 |
Nmr Structure Of D130I Mutant T3-I2, A 32 Residue Peptide From The Alpha 2A Adrenergic Receptor
|
 |
Nmr Structure Of T3-I2, A 32 Residue Peptide From The Alpha-2A Adrenergic Receptor
|
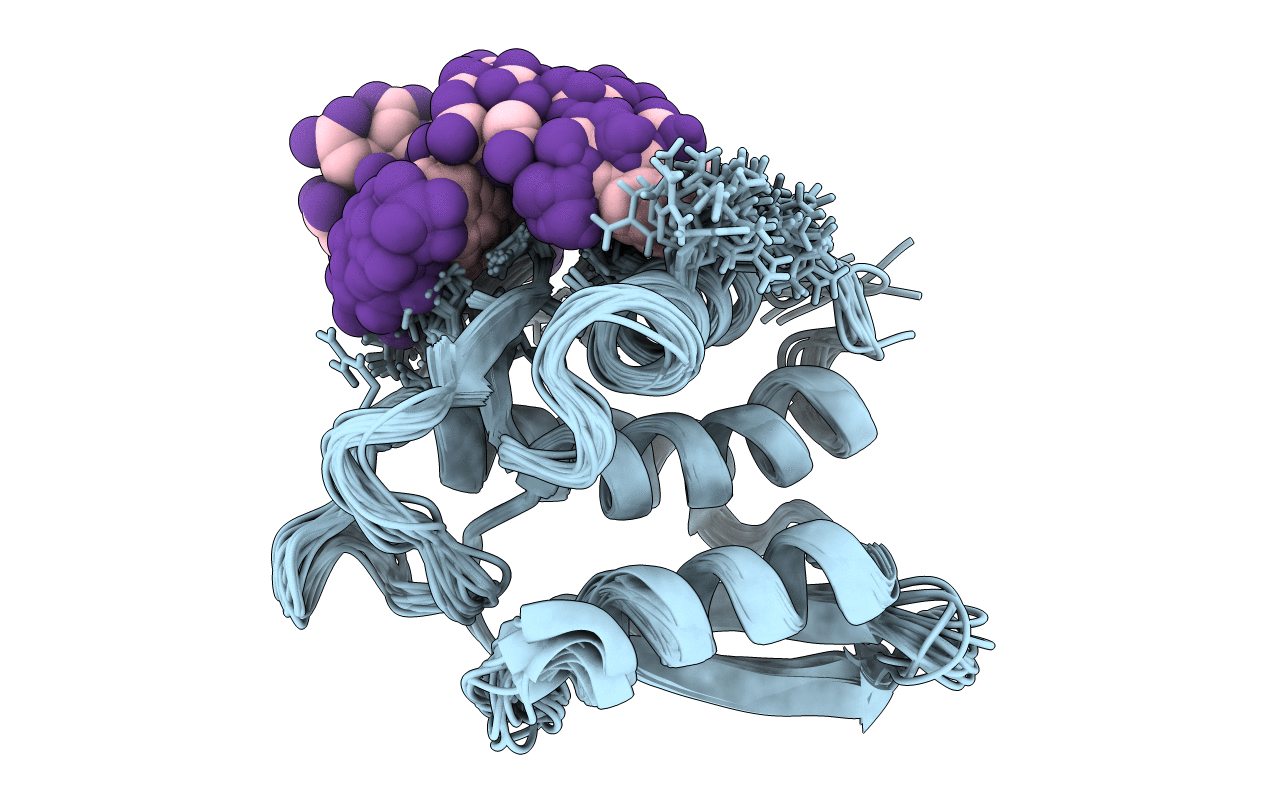 |
Nmr Structure Of The Type Iii Secretory Domain Of Yersinia Yoph Complexed With The Skap-Hom Phospho-Peptide N-Acetyl-Depyddpf-Nh2
Organism: Yersinia pseudotuberculosis
Method: SOLUTION NMR Release Date: 2002-07-24 Classification: HYDROLASE |
 |
Nmr Structure Of T3-I2, A 32 Residue Peptide From The Alpha-2A Adrenergic Receptor
|
 |
Crystal Structure Of The Type Iii Secretory Domain Of Yersinia Yoph Reveals A Domain-Swapped Dimer
Organism: Yersinia pseudotuberculosis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2001-11-28 Classification: HYDROLASE |
 |
 |
High Resolution Solution Structure Of The Heat Shock Cognate-70 Kd Substrate Binding Domain Obtained By Multidimensional Nmr Techniques
Organism: Rattus norvegicus
Method: SOLUTION NMR Release Date: 1999-05-10 Classification: MOLECULAR CHAPERONE |
 |
High Resolution Solution Structure Of The Heat Shock Cognate-70 Kd Substrate Binding Domain Obtained By Multidimensional Nmr Techniques
|
 |
Nmr Structure Of The Substrate Binding Domain Of Dnak, Minimized Average Structure
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 1999-03-02 Classification: MOLECULAR CHAPERONE |
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 1999-03-02 Classification: MOLECULAR CHAPERONE |
 |
Stromelysin-1 Catalytic Domain With Hydrophobic Inhibitor Bound, Ph 7.0, 32Oc, 20 Mm Cacl2, 15% Acetonitrile; Nmr Ensemble Of 20 Structures
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 1996-03-08 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, CA, 0DS |
 |
Stromelysin-1 Catalytic Domain With Hydrophobic Inhibitor Bound, Ph 7.0, 32Oc, 20 Mm Cacl2, 15% Acetonitrile; Nmr Average Of 20 Structures Minimized With Restraints
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 1996-03-08 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: ZN, CA, 0DS |

