Search Count: 35
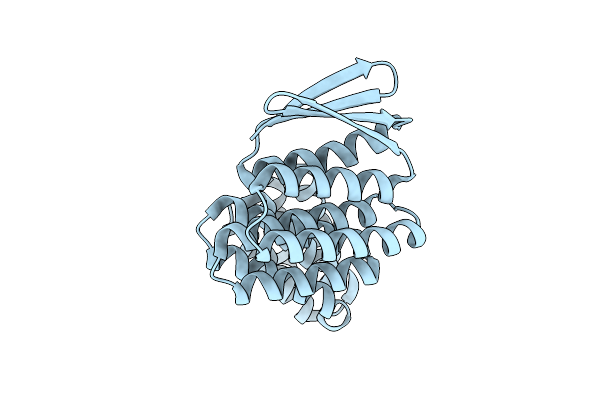 |
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As1
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN Ligands: CL |
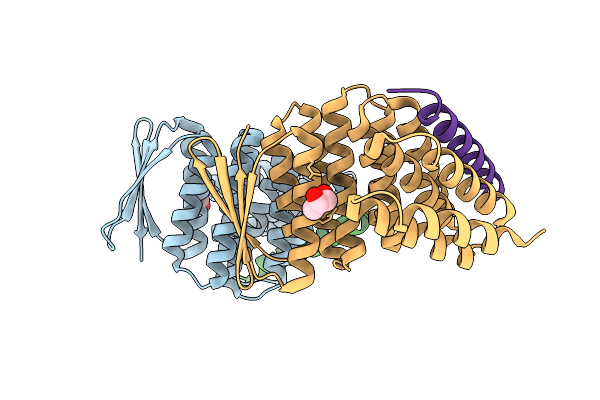 |
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As1 In Complex State He
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN Ligands: PGO |
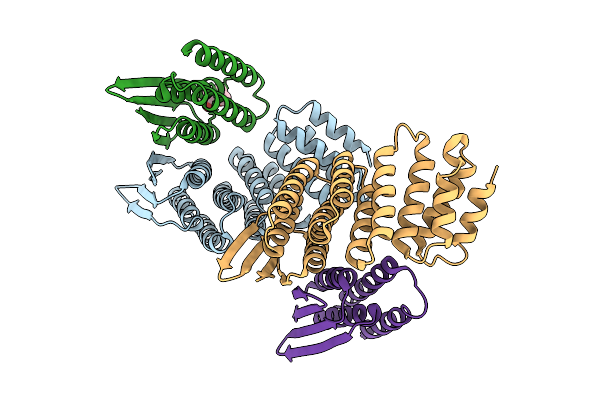 |
Designed Allosteric Facilitated Dissociation Switch As1 In Complex State Th With Methylated Lysines, Crystal #1
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN Ligands: MPD |
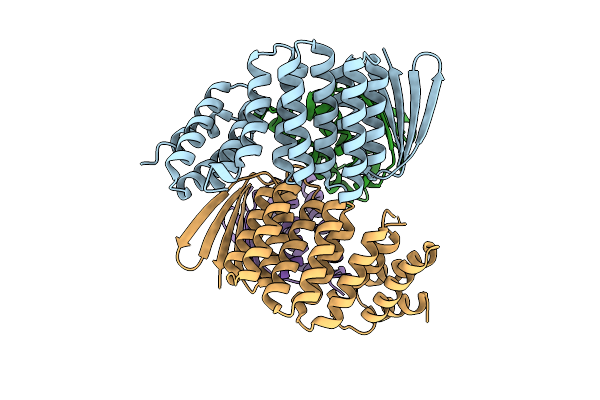 |
Designed Allosteric Facilitated Dissociation Switch As1 In Complex State Th With Methylated Lysines, Crystal #2
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.88 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Designed Allosteric Facilitated Dissociation Switch As1 In Complex State The With Methylated Lysines
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As5
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Designed Allosteric Facilitated Dissociation Switch As5 In Complex State He
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Designed Facilitated Dissociation Target Lhd101An1: Lhd101A With An N-Terminal Extension
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Crystal Structure Of Designed Conformational Switch Effector Peptide Cs221B
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Designed Allosteric Facilitated Dissociation Switch As1_K46L_E50W_K172W_E173Y In Complex State The
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.48 Å Release Date: 2025-08-20 Classification: DE NOVO PROTEIN |
 |
Cryo-Em Structure Of The Pancreatic Atp-Sensitive Potassium Channel Bound To Atp And Repaglinide With Kir6.2-Ctd In The Up Conformation
Organism: Rattus norvegicus, Cricetus cricetus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: MEMBRANE PROTEIN Ligands: ATP, K, POV, P5S, 65I, BJX |
 |
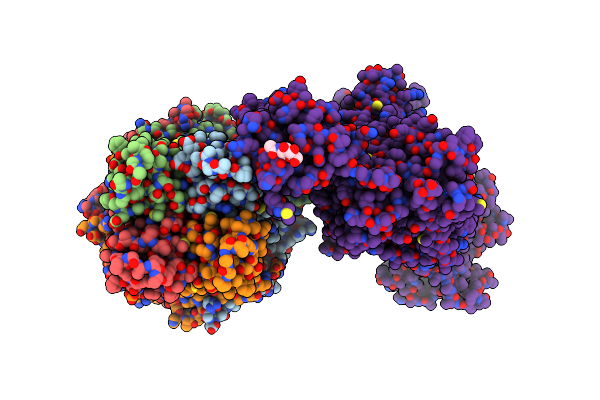
Cryo-Em Structure Of The Pancreatic Atp-Sensitive Potassium Channel Bound To Atp And Repaglinide With Kir6.2-Ctd In The Down Conformation
Organism: Rattus norvegicus, Cricetus cricetus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: MEMBRANE PROTEIN Ligands: ATP, K, POV, P5S, PTY, BJX |
 |
Cryoem Structure Of The Pancreatic Atp-Sensitive Potassium Channel Bound To Atp With Kir6.2-Ctd In The Down Conformation
Organism: Rattus norvegicus, Cricetus cricetus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: MEMBRANE PROTEIN Ligands: ATP, NAG |
 |
Cryo-Em Structure Of The Pancreatic Atp-Sensitive Potassium Channel Bound To Atp And Repaglinide With Sur1-In Conformation
Organism: Rattus norvegicus, Cricetus cricetus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: MEMBRANE PROTEIN Ligands: ATP, K, P5S, POV, PTY, BJX |
 |
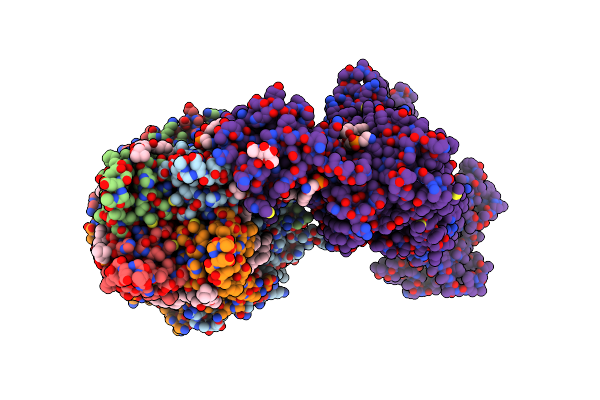
Cryo-Em Structure Of The Pancreatic Atp-Sensitive Potassium Channel Bound To Atp And Repaglinide With Sur1-Out Conformation
Organism: Rattus norvegicus, Cricetus cricetus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: MEMBRANE PROTEIN Ligands: ATP, K, POV, P5S, PTY, BJX |
 |
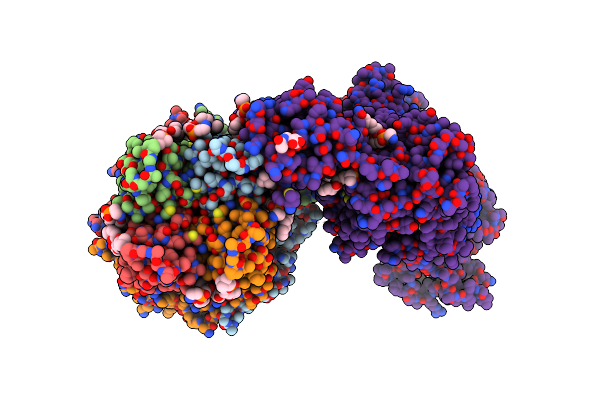
Cryo-Em Structure Of The Pancreatic Atp-Sensitive Potassium Channel Bound To Atp And Glibenclamide With Kir6.2-Ctd In The Up Conformation
Organism: Rattus norvegicus, Cricetus cricetus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: MEMBRANE PROTEIN Ligands: ATP, PTY, POV, P5S, GBM, NAG |
 |
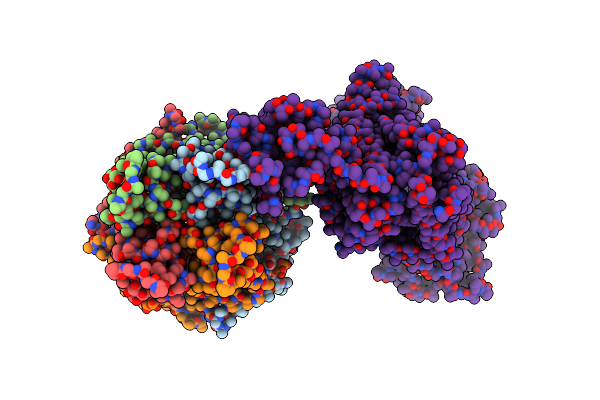
Cryo-Em Structure Of The Pancreatic Atp-Sensitive Potassium Channel In The Presence Of Carbamazepine And Atp With Kir6.2-Ctd In The Down Conformation
Organism: Rattus norvegicus, Cricetus cricetus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: MEMBRANE PROTEIN Ligands: PTY, POV, P5S, ATP, NAG |
 |
Cryo-Em Structure Of The Pancreatic Atp-Sensitive Potassium Channel In The Presence Of Glibenclamide And Atp With Kir6.2-Ctd In The Down Conformation
Organism: Rattus norvegicus, Cricetus cricetus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: MEMBRANE PROTEIN Ligands: ATP |
 |
Cryo-Em Structure Of The Pancreatic Atp-Sensitive Potassium Channel In The Presence Of Carbamazepine And Atp With Kir6.2-Ctd In The Up Conformation
Organism: Rattus norvegicus, Cricetus cricetus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: MEMBRANE PROTEIN Ligands: ATP |
 |
Cryoem Structure Of The Pancreatic Atp-Sensitive Potassium Channel In The Atp-Bound State With Kir6.2-Ctd In The Up Conformation
Organism: Rattus norvegicus, Cricetus cricetus
Method: ELECTRON MICROSCOPY Release Date: 2022-08-31 Classification: MEMBRANE PROTEIN Ligands: ATP |

