Search Count: 158
 |
Organism: Entacmaea quadricolor
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: FLUORESCENT PROTEIN Ligands: EDO |
 |
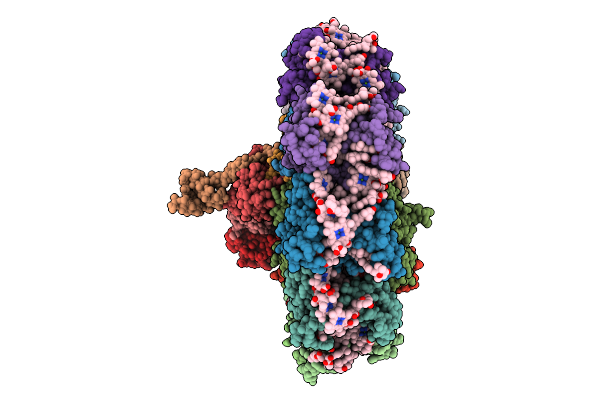
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Tribonema minus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: PHOTOSYNTHESIS Ligands: CLA, BCR, DD6, LMT, LHG, A1L1G, LMG, PQN, SF4 |
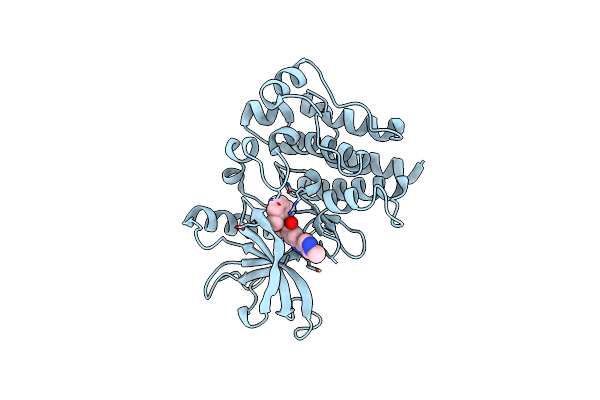 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: CELL CYCLE Ligands: A1A6M, CL, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2025-02-19 Classification: TRANSFERASE/INHIBITOR Ligands: A1AZP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-02-19 Classification: TRANSFERASE/INHIBITOR Ligands: A1AZ8, MG, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-02-12 Classification: HYDROLASE Ligands: ZN, MG, A1D8N |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-02-12 Classification: HYDROLASE Ligands: ZN, MG, A1D8O |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-02-12 Classification: HYDROLASE Ligands: ZN, MG, A1D8P |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-02-12 Classification: HYDROLASE Ligands: ZN, MG, A1D8Q |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2025-01-29 Classification: HYDROLASE Ligands: A1EIY, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-01-29 Classification: HYDROLASE Ligands: A1EIZ |
 |
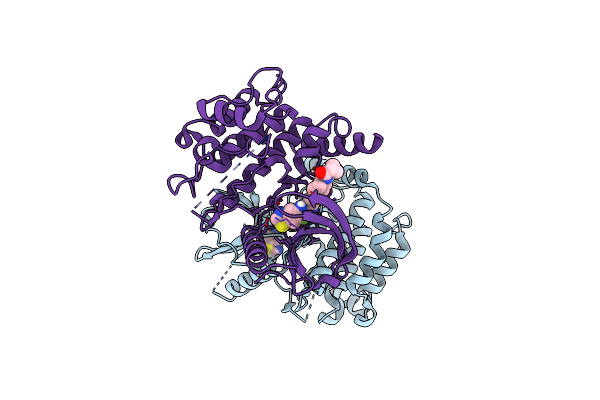
Organism: Fusarium tricinctum
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2024-12-11 Classification: TRANSFERASE Ligands: PPV, TOE, MOE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2024-11-13 Classification: IMMUNE SYSTEM Ligands: 6IL |
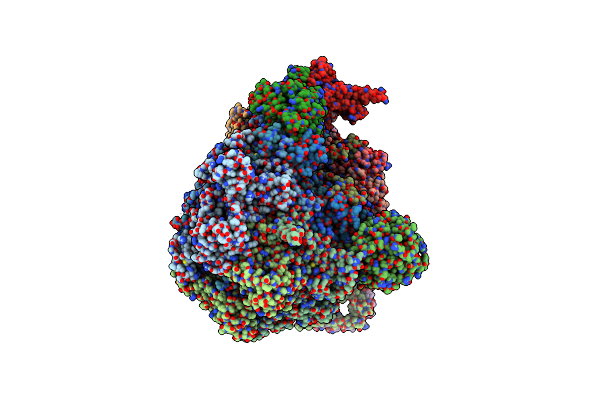 |
Cryo-Em Structure Of The Ycf2-Ftshi Motor Complex From Chlamydomonas Reinhardtii In Amppnp Bound State
Organism: Chlamydomonas reinhardtii
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: MEMBRANE PROTEIN Ligands: LMG, ANP, MG, SQD, DGA, Y01, A1LXL, DGD, ZN |
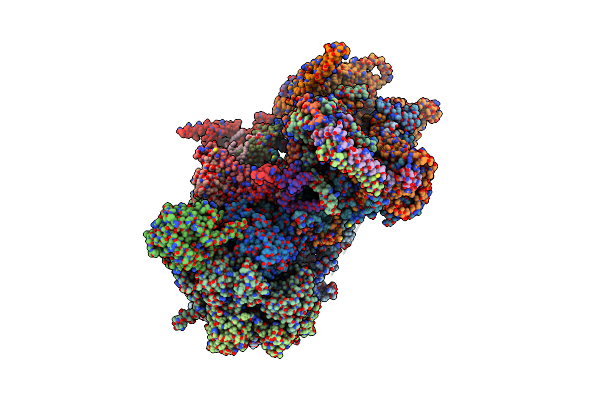 |
Cryo-Em Structure Of The Ycf2-Ftshi Motor Complex From Chlamydomonas Reinhardtii In Apo State
Organism: Chlamydomonas reinhardtii
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: MEMBRANE PROTEIN Ligands: LMG, MG, SQD, DGA, Y01, DGD, A1LXL, ZN |

