Search Count: 27
 |
Proton Conductance By Human Uncoupling Protein 1 Is Inhibited By Both Purine And Pyrimidine Nucleotides
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: TRANSPORT PROTEIN Ligands: UTP, CDL |
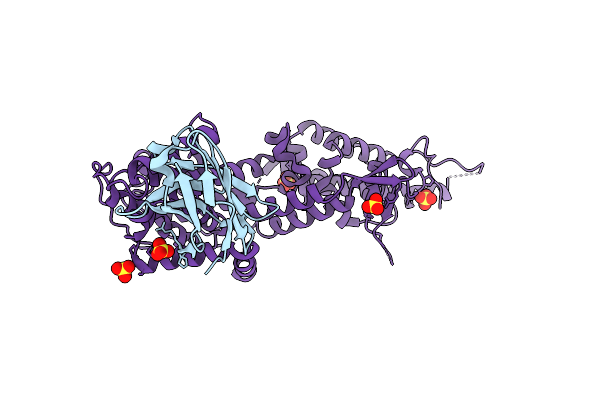 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-11-01 Classification: SIGNALING PROTEIN Ligands: SO4 |
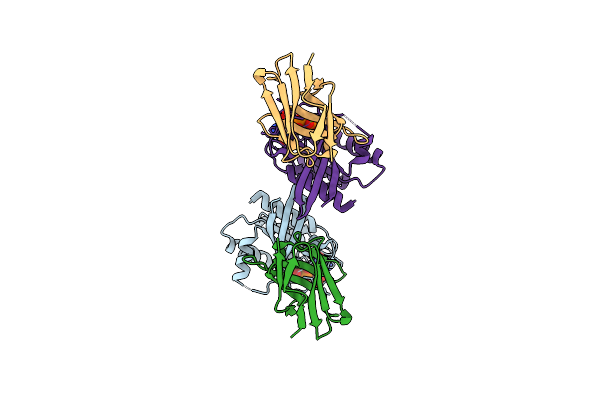 |
Organism: Lama glama, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-11-01 Classification: SIGNALING PROTEIN Ligands: MG, GDP |
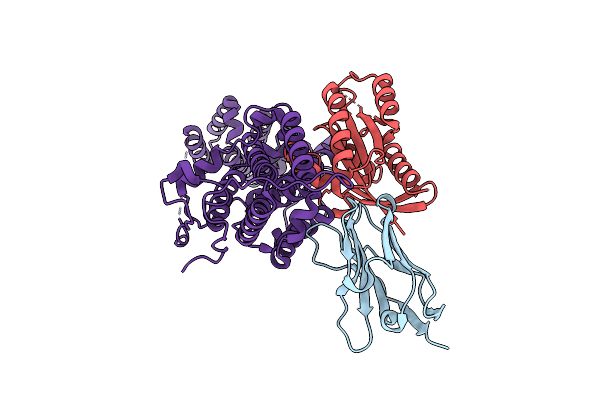 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-11-01 Classification: SIGNALING PROTEIN |
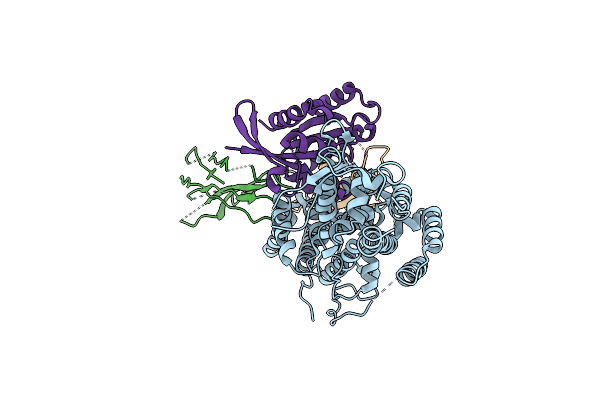 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:3.13 Å Release Date: 2023-11-01 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct, Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2023-06-07 Classification: MEMBRANE PROTEIN Ligands: GTP, CDL |
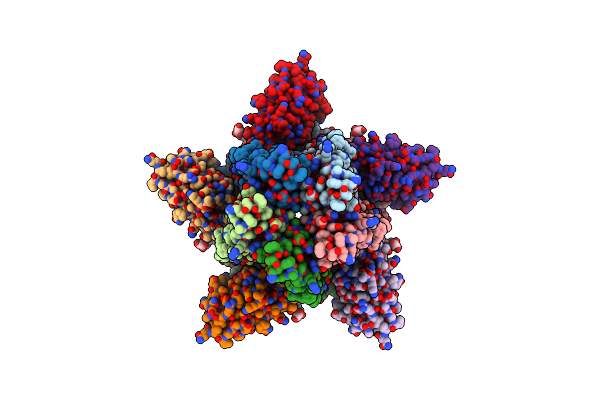 |
Cryoem Structure Of A Beta3K279T Gaba(A)R Homomer In Complex With Histamine And Megabody Mb25
Organism: Homo sapiens, Helicobacter pylori (strain g27), Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2021-08-04 Classification: MEMBRANE PROTEIN Ligands: NAG, HSM |
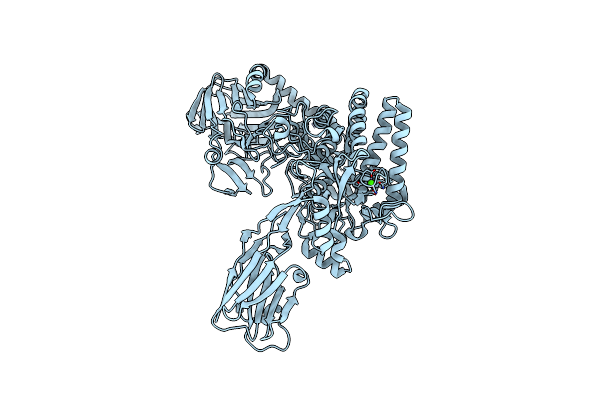 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-01-13 Classification: STRUCTURAL PROTEIN Ligands: CA |
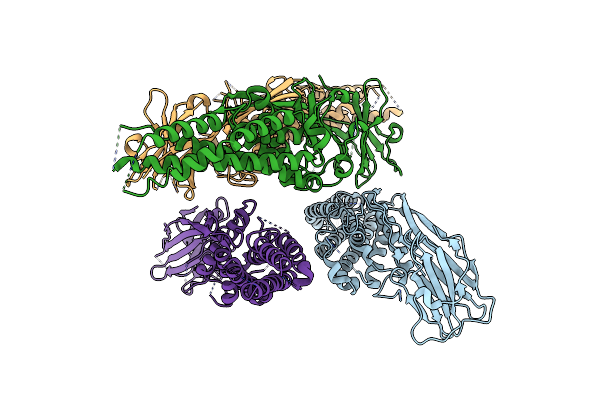 |
Organism: Lama glama, Helicobacter pylori (strain g27)
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2021-01-13 Classification: STRUCTURAL PROTEIN |
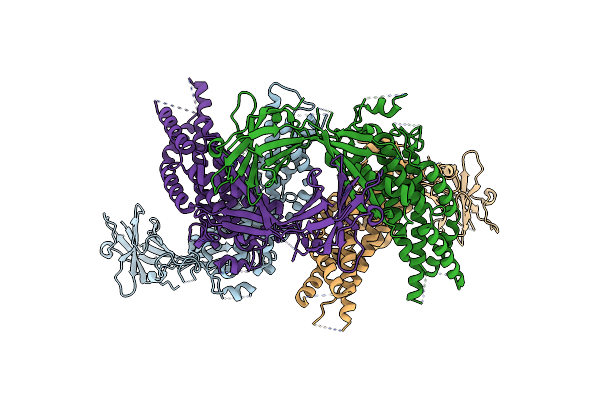 |
Organism: Helicobacter pylori (strain g27), Helicobacter pylori g27
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-01-13 Classification: STRUCTURAL PROTEIN |
 |
Molecular Scaffolds Expand The Nanobody Toolkit For Cryo-Em Applications: Crystal Structure Of Mb-Chopq-Nb207
Organism: Helicobacter pylori, Helicobacter pylori (strain g27)
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2019-12-18 Classification: STRUCTURAL PROTEIN Ligands: CL |
 |
Organism: Thermothelomyces thermophila (strain atcc 42464 / bcrc 31852 / dsm 1799), Lama glama
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2019-01-09 Classification: MEMBRANE PROTEIN Ligands: BKC, CDL, P6G |
 |
Organism: Homo sapiens, Camelus dromedarius, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-12-07 Classification: Hydrolase/Antibody Ligands: NAG, CA, NA |
 |
Organism: Camelus dromedarius
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2016-12-07 Classification: IMMUNE SYSTEM Ligands: CL |
 |
Organism: Pestivirus strain d32/00_hobi
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: SGM, CL |
 |
Crystal Structure Of Substrate-Like, Unprocessed N-Terminal Protease Npro Mutant S169P
Organism: Pestivirus strain d32/00_hobi
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: SGM, CL, OH |
 |
Crystal Structure Of Product-Like, Processed N-Terminal Protease Npro With Internal His-Tag
Organism: Pestivirus strain d32/00_hobi
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: SGM, CL |
 |
Crystal Structure Of Product-Like, Processed N-Terminal Protease Npro With Mercury
Organism: Pestivirus strain d32/00_hobi
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: SGM, HG |
 |
Crystal Structure Of Product-Like, Processed N-Terminal Protease Npro With Iridium
Organism: Pestivirus strain d32/00_hobi
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: SGM, OH, IR3 |
 |
Crystal Structure Of Product-Like, Processed N-Terminal Protease Npro At Ph 3
Organism: Pestivirus strain d32/00_hobi
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-05-15 Classification: HYDROLASE Ligands: SGM, CL |

