Search Count: 21
 |
Organism: Gallus gallus
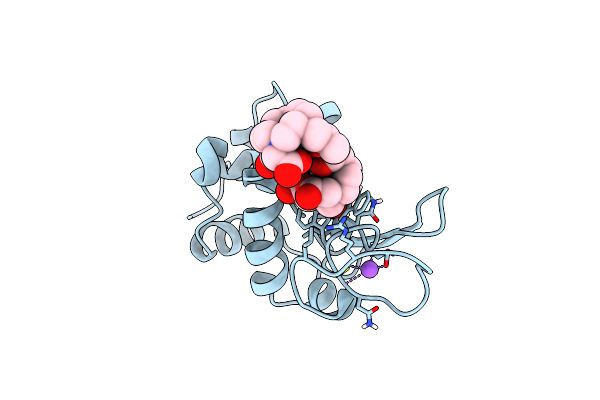
Method: X-RAY DIFFRACTION Resolution:0.80 Å Release Date: 2025-05-28 Classification: HYDROLASE Ligands: GD, DO3, NO3, CL, NA |
 |
Organism: Gallus gallus
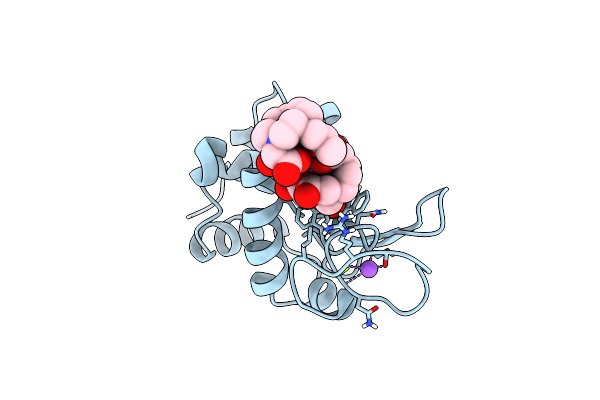
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2025-05-28 Classification: HYDROLASE Ligands: GD, DO3, NO3, CL, NA |
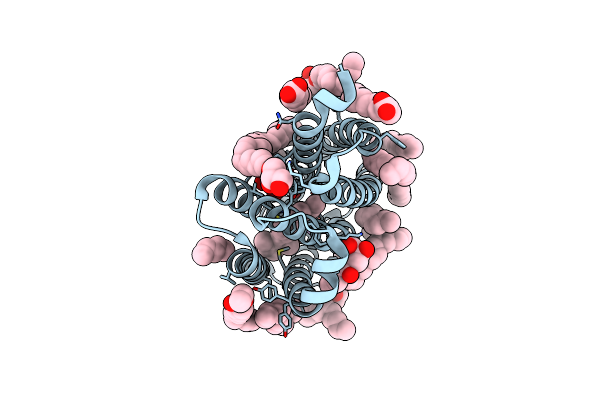 |
True-Atomic Resolution Crystal Structure Of The Closed State Of The Viral Channelrhodopsin Olpvr1
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN |
 |
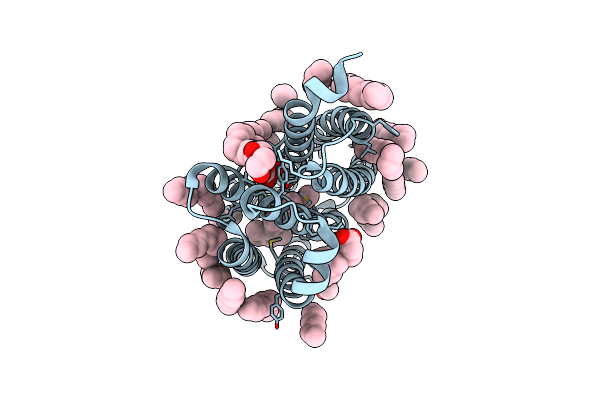
True-Atomic Resolution Crystal Structure Of The Open State Of The Viral Channelrhodopsin Olpvr1
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.34 Å Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN Ligands: RET, NA, 97N, OLC, LFA, GOL |
 |
Structure Of The Viral Channelrhodopsin Olpvr1 At Low Ph Obtained By Soaking
Organism: Organic lake phycodnavirus
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN Ligands: RET, 97N, LFA, OLC |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2024-11-06 Classification: IMMUNE SYSTEM Ligands: GOL, PGE |
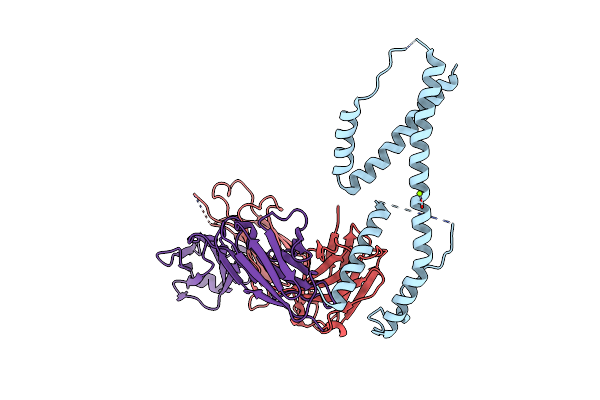 |
Crystal Structure Of The Fab Fragment Of The Anti-Il-6 Antibody I9H In Complex With A Domain-Swapped Il-6 Dimer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2024-11-06 Classification: IMMUNE SYSTEM Ligands: MG |
 |
Crystal Structure Of The Fab Fragment Of The Anti-Il-6 Antibody 68F2 In Complex With A Domain-Swapped Il-6 Dimer
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Resolution:2.93 Å Release Date: 2024-11-06 Classification: IMMUNE SYSTEM Ligands: SO4 |
 |
Organism: Hirudo medicinalis
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2023-02-08 Classification: HYDROLASE |
 |
Organism: Hirudo medicinalis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-02-08 Classification: HYDROLASE Ligands: GOL, CL |
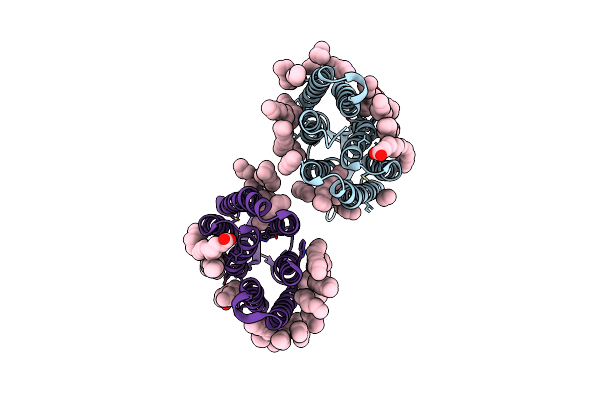 |
Crystal Structure Of A Light-Driven Proton Pump Lr (Mac) From Leptosphaeria Maculans
Organism: Leptosphaeria maculans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-07-07 Classification: MEMBRANE PROTEIN Ligands: LFA, OLA |
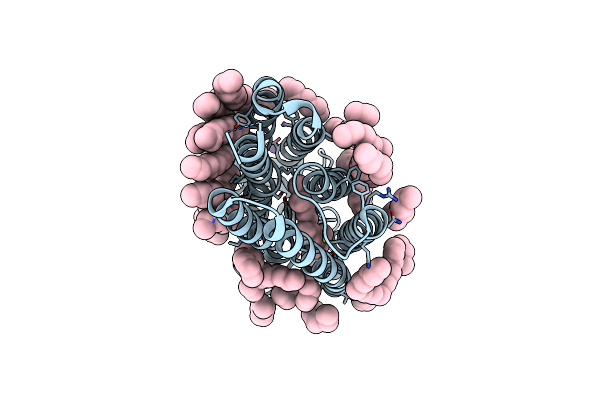 |
Crystal Structure Of The D116N Mutant Of The Light-Driven Sodium Pump Kr2 In The Monomeric Form, Ph 4.6
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-04-08 Classification: MEMBRANE PROTEIN |
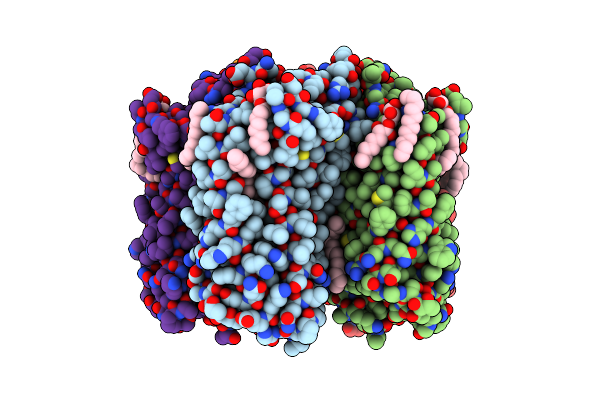 |
Crystal Structure Of The D116N Mutant Of The Light-Driven Sodium Pump Kr2 In The Pentameric Form, Ph 8.0
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-04-08 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of The Steady-State-Smx Activated State Of The Light-Driven Sodium Pump Kr2 In The Pentameric Form At Room Temperature, Ph 8.0
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-04-08 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, NA, RET, GOL, OLA |
 |
Crystal Structure Of The H30A Mutant Of The Light-Driven Sodium Pump Kr2 In The Pentameric Form, Ph 8.0
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-04-08 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, NA, BOG, RET |
 |
Crystal Structure Of The Light-Driven Sodium Pump Kr2 In The Pentameric Form At Room Temperature, Ph 8.0
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-04-08 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of The Light-Driven Sodium Pump Kr2 In The Pentameric Form, Ph 8.0
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-04-08 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, BOG, RET, NA |
 |
Crystal Structure Of The Steady-State Activated State Of The Light-Driven Sodium Pump Kr2 In The Pentameric Form At Room Temperature, Ph 8.0
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-04-08 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, NA, RET, GOL, OLA |
 |
Crystal Structure Of The O-State Of The Light-Driven Sodium Pump Kr2 In The Pentameric Form, Ph 8.0
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-02-12 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, NA, BOG, OLA, GOL |
 |
Structure Of Bacteriorhodopsin From Crystals Grown At 20 Deg Celcius Using Glyncoc15+4 As An Lcp Host Lipid
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2017-07-12 Classification: MEMBRANE PROTEIN |

