Search Count: 24
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: RECOMBINATION |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: RECOMBINATION |
 |
Ms2 Bacteriophage Coat Protein After Reassembly As A Nanocrate (Ms2Nc) With No Cargo And With Waters Modeled
Organism: Escherichia phage ms2
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: VIRUS |
 |
Organism: Escherichia phage ms2
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: VIRUS |
 |
Crystal Structure Of E. Coli Class Ia Ribonucleotide Reductase Alpha Subunit Bound To Two Atp Molecules
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2024-09-04 Classification: OXIDOREDUCTASE Ligands: ATP, MG, GOL, MES |
 |
Crystal Structure Of E. Coli Class Ia Ribonucleotide Reductase Alpha Subunit Bound To Datp
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2024-09-04 Classification: OXIDOREDUCTASE Ligands: DTP, MG, MES, GOL |
 |
Crystal Structure Of E. Coli Class Ia Ribonucleotide Reductase Alpha Subunit W28A Variant Bound To Cdp And Two Molecules Of Atp
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2024-09-04 Classification: OXIDOREDUCTASE Ligands: SO4, ATP, MG, CDP |
 |
Crystal Structure Of E. Coli Class Ia Ribonucleotide Reductase Alpha Subunit W28A Variant Bound To Datp And Atp
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2024-09-04 Classification: OXIDOREDUCTASE Ligands: DTP, MG, ATP |
 |
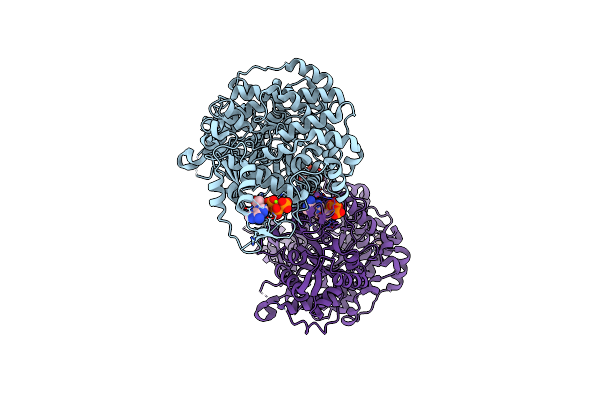
Crystal Structure Of E. Coli Class Ia Ribonucleotide Reductase Alpha Subunit W28A Variant Bound To Datp And Gtp
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:3.55 Å Release Date: 2024-09-04 Classification: OXIDOREDUCTASE Ligands: DTP, MG, GTP |
 |
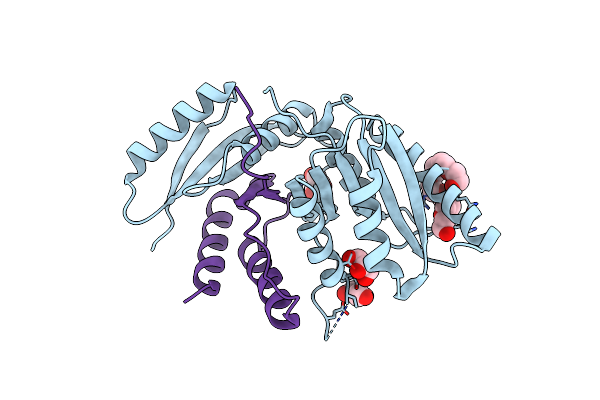
Crystal Structure Of Datp Bound E. Coli Class Ia Ribonucleotide Reductase Alpha Construct Fused With The C-Terminal Tail Of E. Coli Class Ia Beta Subunit
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-09-04 Classification: OXIDOREDUCTASE Ligands: DTP, MG, NA, CL, GOL |
 |
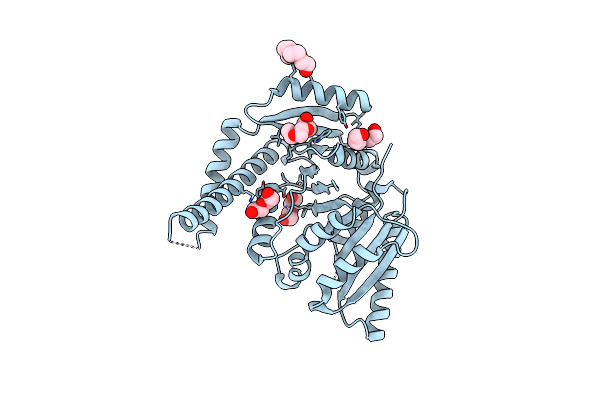
C-Terminal Regulatory Domain Of The Chloride Transporter Kcc-1 From C. Elegans, Proteolyzed During Crystallization
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-07-15 Classification: TRANSPORT PROTEIN Ligands: P33, GOL |
 |
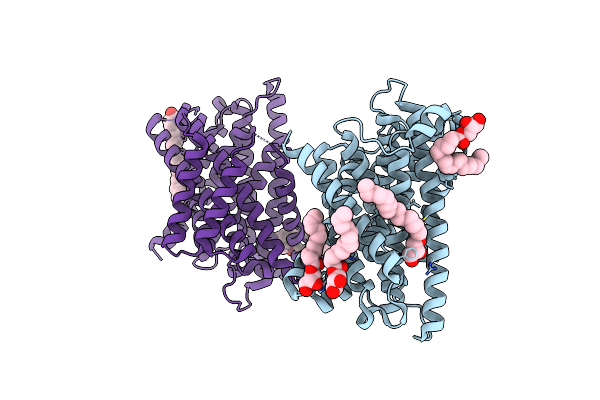
C-Terminal Regulatory Domain Of The Chloride Transporter Kcc-1 From C. Elegans
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-07-15 Classification: TRANSPORT PROTEIN Ligands: PGE, PEG, PG4 |
 |
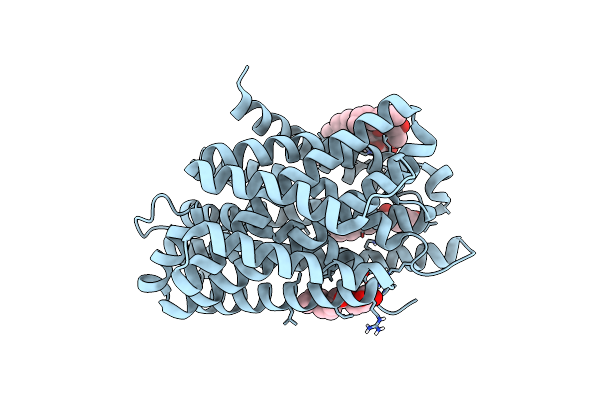
Crystal Structure Of The Deinococcus Radiodurans Nramp/Mnth Divalent Transition Metal Transporter G45R Mutant In An Inward Occluded State
Organism: Deinococcus radiodurans r1
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2019-02-13 Classification: TRANSPORT PROTEIN Ligands: OLC |
 |
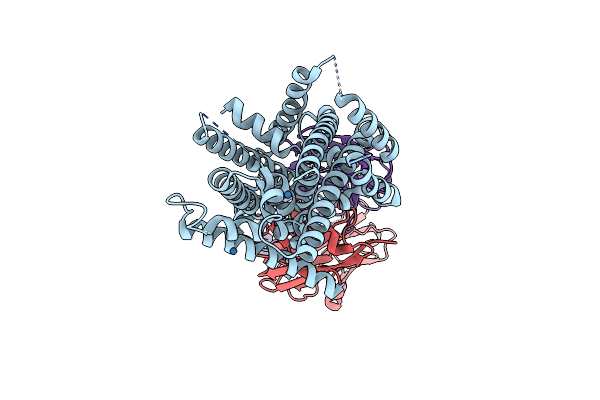
Crystal Structure Of The Deinococcus Radiodurans Nramp/Mnth Divalent Transition Metal Transporter In The Outward-Open, Apo Conformation
Organism: Deinococcus radiodurans r1
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2019-02-13 Classification: TRANSPORT PROTEIN Ligands: OLC |
 |
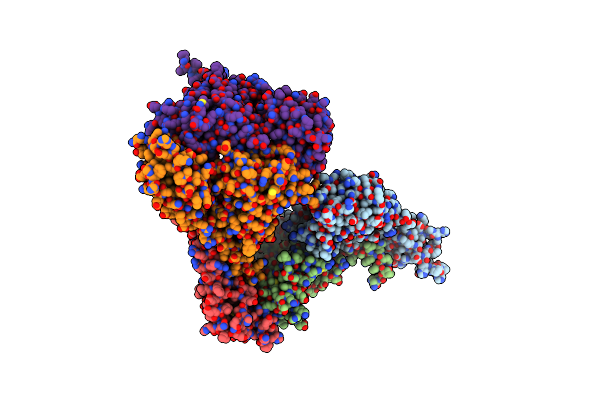
Crystal Structure Of Deinococcus Radiodurans Mnth, An Nramp-Family Transition Metal Transporter, In The Inward-Open Apo State
Organism: Deinococcus radiodurans, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.94 Å Release Date: 2019-02-13 Classification: TRANSPORT PROTEIN Ligands: OS |
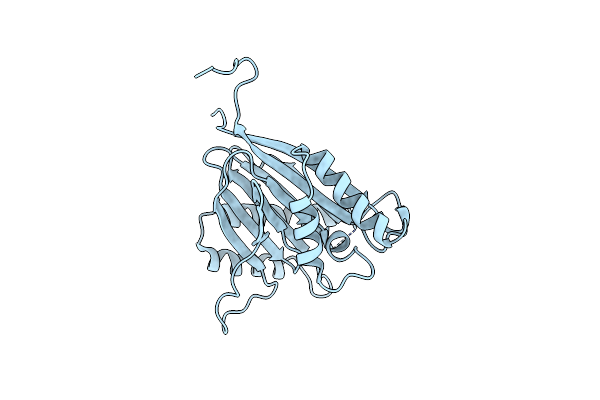 |
Structure Of The Phosphatase Domain Of The Cell Fate Determinant Spoiie From Bacillus Subtilis In A Crystal Form Without Domain Swapping
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2017-05-31 Classification: TRANSFERASE |
 |
Structure Of The Pp2C Phosphatase Domain And A Fragment Of The Regulatory Domain Of The Cell Fate Determinant Spoiie From Bacillus Subtilis
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:3.91 Å Release Date: 2017-05-31 Classification: HYDROLASE |
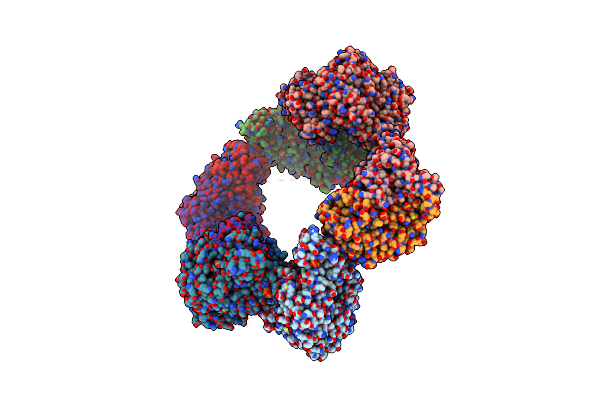 |
Crystal Structure Of The Datp Inhibited E. Coli Class Ia Ribonucleotide Reductase Complex Bound To Cdp And Datp At 2.97 Angstroms Resolution
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2016-01-20 Classification: OXIDOREDUCTASE Ligands: CDP, DAT, MG, DTP, FEO |
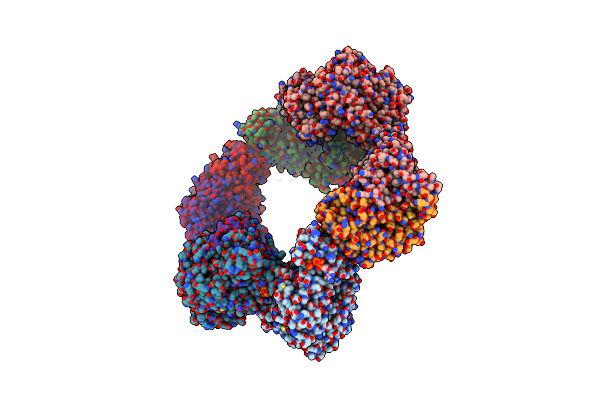 |
Crystal Structure Of The Datp Inhibited E. Coli Class Ia Ribonucleotide Reductase Complex Bound To Udp And Datp At 3.25 Angstroms Resolution
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2016-01-20 Classification: OXIDOREDUCTASE Ligands: UDP, DTP, MG, FEO |
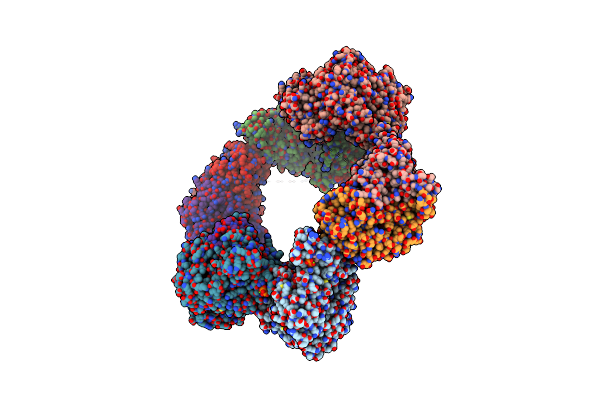 |
Crystal Structure Of The Datp Inhibited E. Coli Class Ia Ribonucleotide Reductase Complex Bound To Adp And Dgtp At 3.40 Angstroms Resolution
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2016-01-20 Classification: OXIDOREDUCTASE Ligands: DGT, MG, ADP, DAT, FEO |

