Search Count: 10
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2015-12-09 Classification: OXIDOREDUCTASE |
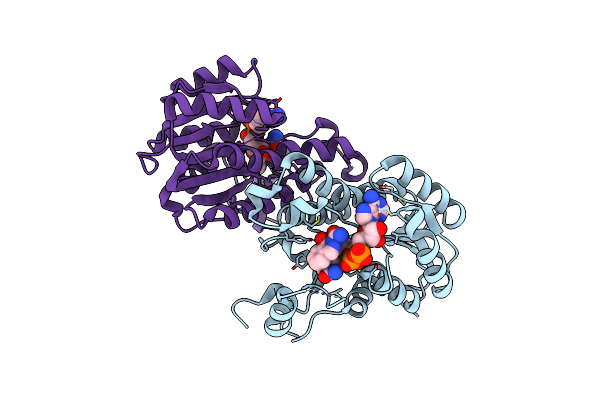 |
Crystal Structure Of E. Coli Fabi Bound To The Carbamoylated Benzodiazaborine Inhibitor 14B.
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2015-12-09 Classification: Oxidoreductase/Oxidoreductase Inhibitor Ligands: NAD, BBN |
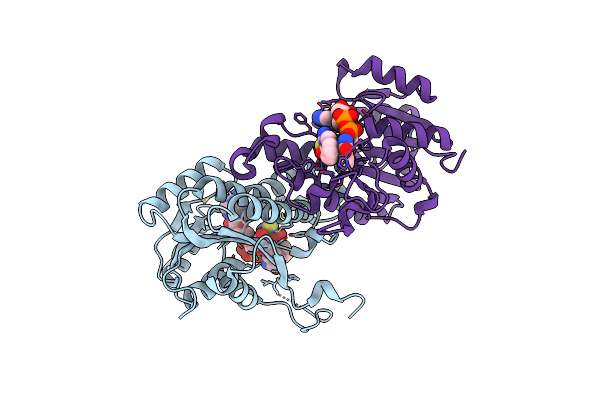 |
Crystal Structure Of E. Coli Fabi Bound To The Thiocarbamoylated Benzodiazaborine Inhibitor 35B.
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2015-12-09 Classification: Oxidoreductase/Oxidoreductase Inhibitor Ligands: NAD, CJ3 |
 |
Crystal Structure Of The Glutaminyl-Trna Synthetase From Deinococcus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-04-24 Classification: LIGASE |
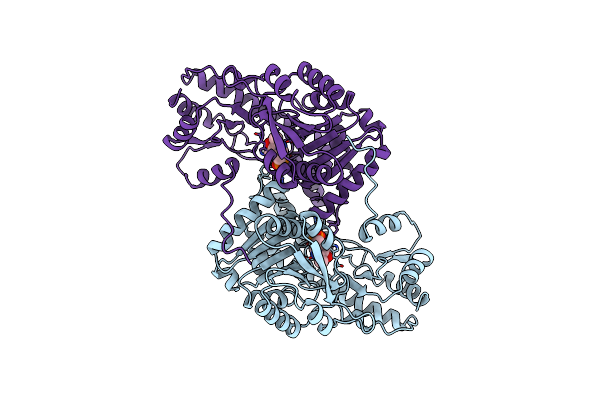 |
Structural Basis For The Catalytic Activity Of Aspartate Aminotransferase K258H Lacking The Pyridoxal-5'-Phosphate Binding Lysine Residue
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1994-10-15 Classification: TRANSFERASE(AMINOTRANSFERASE) Ligands: PMP |
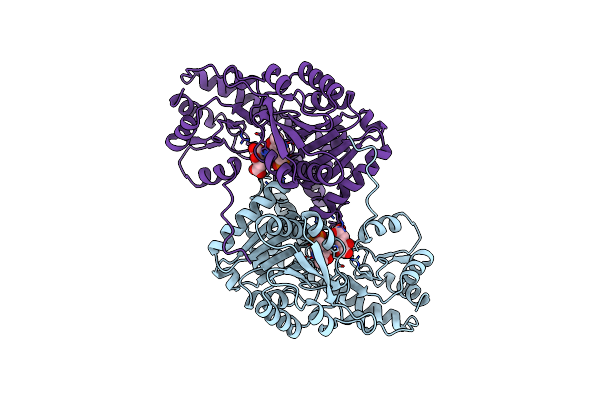 |
Structural Basis For The Catalytic Activity Of Aspartate Aminotransferase K258H Lacking The Pyridoxal-5'-Phosphate Binding Lysine Residue
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 1994-10-15 Classification: TRANSFERASE(AMINOTRANSFERASE) Ligands: PMP, AKG |
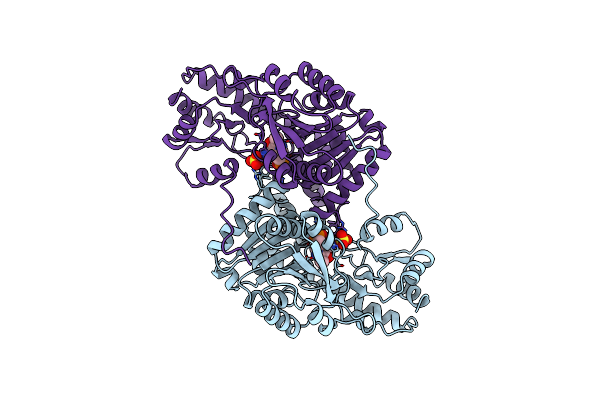 |
Structural Basis For The Catalytic Activity Of Aspartate Aminotransferase K258H Lacking The Pyridoxal-5'-Phosphate Binding Lysine Residue
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 1994-10-15 Classification: TRANSFERASE(AMINOTRANSFERASE) Ligands: SO4, PMP |
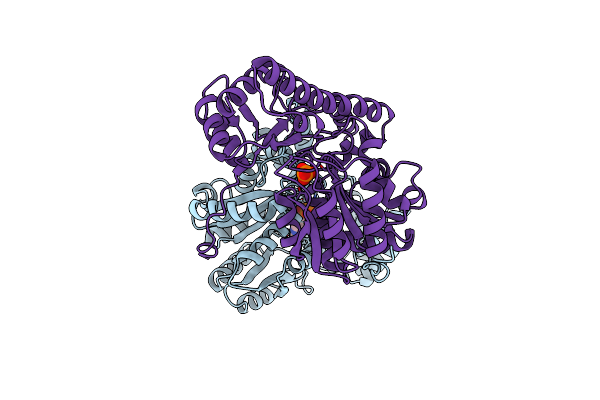 |
Structural Basis For The Catalytic Activity Of Aspartate Aminotransferase K258H Lacking Its Pyridoxal-5'-Phosphate-Binding Lysine Residue
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1994-07-31 Classification: TRANSFERASE(AMINOTRANSFERASE) Ligands: PLP, PO4 |
 |
Structural Basis For The Catalytic Activity Of Aspartate Aminotransferase K258H Lacking Its Pyridoxal-5'-Phosphate-Binding Lysine Residue
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1994-07-31 Classification: TRANSFERASE(AMINOTRANSFERASE) Ligands: PPD |
 |
Structural Basis For The Catalytic Activity Of Aspartate Aminotransferase K258H Lacking Its Pyridoxal-5'-Phosphate-Binding Lysine Residue
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1994-07-31 Classification: TRANSFERASE(AMINOTRANSFERASE) Ligands: PPE |

