Search Count: 22
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: LIGASE Ligands: EDO, MPD |
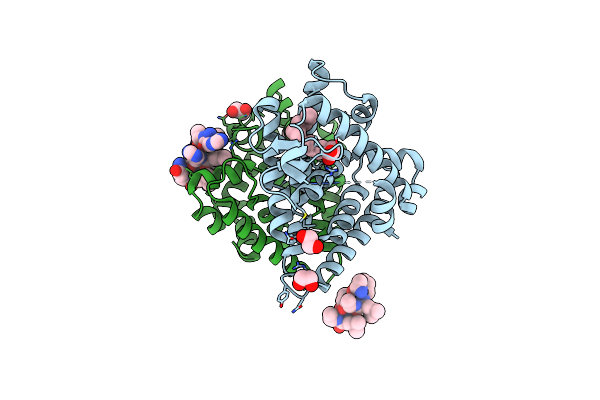 |
Crystal Structure Of Hnf4 Alpha Lbd In Complexes With Palmitic Acid And Grip-1 Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-10 Classification: DNA BINDING PROTEIN Ligands: PLM, EDO |
 |
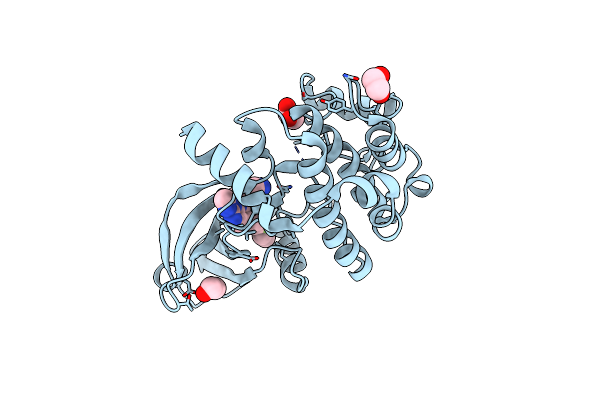
Crystal Structure Of Human Ephrin Type-A Receptor 2 (Epha2) Kinase Domain In Complex With Jg165
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-11-22 Classification: TRANSFERASE Ligands: WT3, EDO |
 |
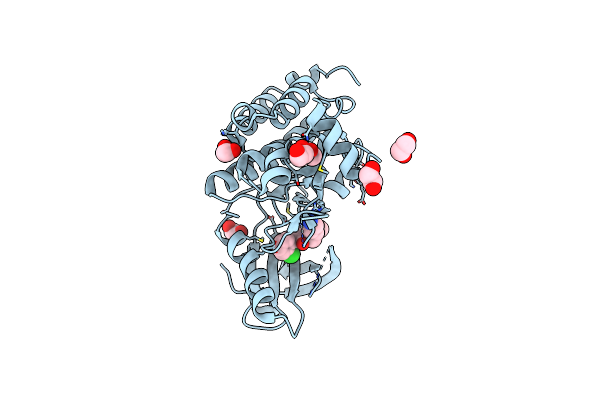
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: SJL, EDO |
 |
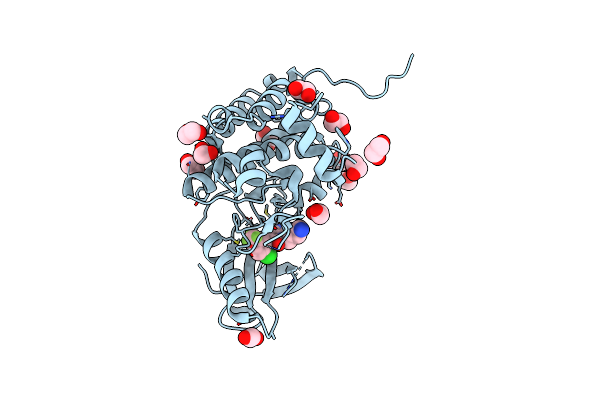
Crystal Structure Of Human Ephrin Type-A Receptor 2 (Epha2) Kinase Domain In Complex With Mr21
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-11-23 Classification: TRANSFERASE Ligands: EDO, QRR |
 |
Crystal Structure Of Human Ephrin Type-A Receptor 2 (Epha2) Kinase Domain In Complex With Mral5
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-11-23 Classification: TRANSFERASE Ligands: EDO, QRD |
 |
Crystal Structure Of Human Ephrin Type-A Receptor 2 (Epha2) Kinase Domain In Complex With Ldn-211904
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-11-16 Classification: TRANSFERASE Ligands: EDO, QJI |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2022-05-04 Classification: LIGASE Ligands: EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-05-04 Classification: LIGASE Ligands: SO4, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-05-04 Classification: LIGASE Ligands: EDO |
 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2022-05-04 Classification: LIGASE Ligands: SO4, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2022-05-04 Classification: LIGASE Ligands: EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2022-05-04 Classification: LIGASE Ligands: SO4, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2022-05-04 Classification: LIGASE Ligands: IOD, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2022-05-04 Classification: LIGASE Ligands: EDO, CL |
 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2022-05-04 Classification: LIGASE Ligands: EDO |
 |
Crystal Structure Of Phosphorylated Pt220 Casein Kinase I Delta (Ck1D), Conformation 1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2022-04-13 Classification: TRANSFERASE Ligands: SO4, EDO, AMP, ADN |
 |
Crystal Structure Of Phosphorylated Pt220 Casein Kinase I Delta (Ck1D), Conformation 2 And 3
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-04-13 Classification: TRANSFERASE Ligands: CIT, EDO, AMP |

