Search Count: 37
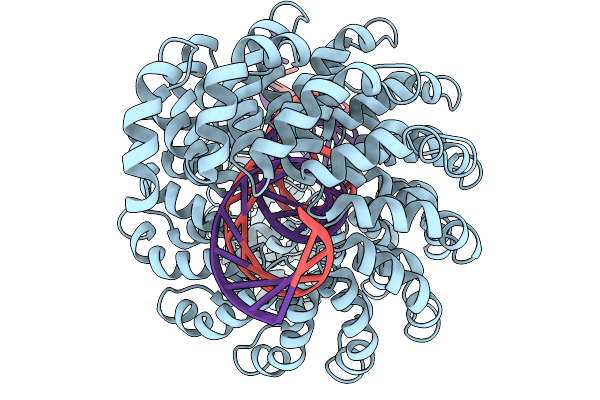 |
Organism: Burkholderia cenocepacia, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
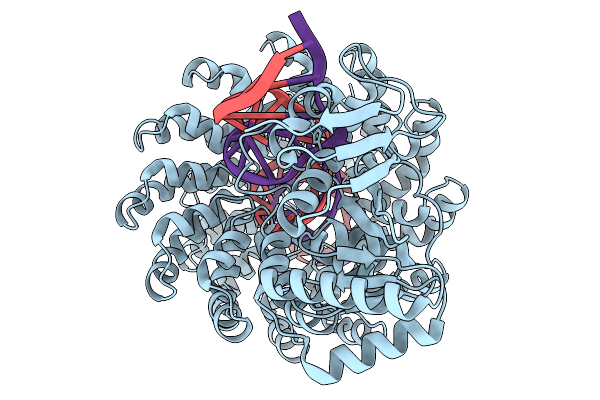 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
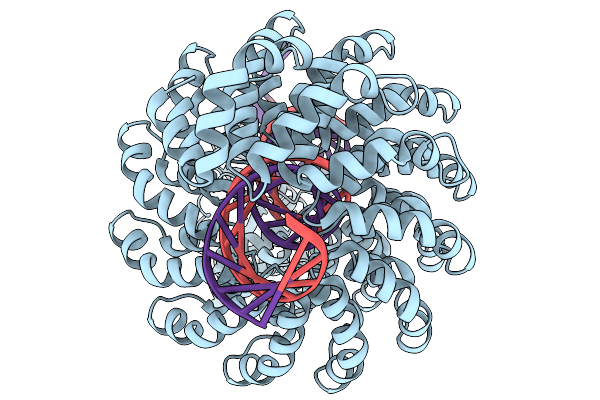 |
Organism: Xanthomonas, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
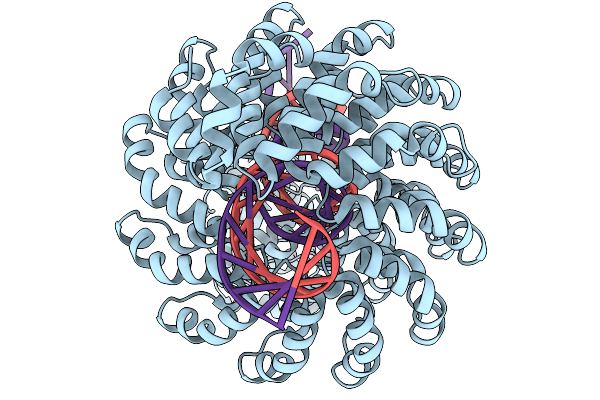 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2025-01-29 Classification: HYDROLASE Ligands: A1EIY, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-01-29 Classification: HYDROLASE Ligands: A1EIZ |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.00 Å Release Date: 2025-01-01 Classification: IMMUNE SYSTEM |
 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2024-02-07 Classification: METAL BINDING PROTEIN Ligands: CA, MN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-12-13 Classification: CELL CYCLE Ligands: XSE |
 |
Local Refined Cryo-Em Structure Of Omicron Ba.5 Rbd In Complex With 8-9D Fab
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Cryo-Em Structure Of Sars-Cov2 Omicron Ba.5 Spike In Complex With 8-9D Fabs
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-11-29 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-07-12 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Sars-Cov-2 Ba.4 Variants S Ectodomain Trimer In Complex With Neutralizing Antibody 10-5B And 6-2C
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Sars-Cov-2 Ba.1 Variants S Ectodomain Trimer In Complex With Neutralizing Antibody 10-5B And 6-2C
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Duganella sp. zlp-xi
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-04-12 Classification: FLAVOPROTEIN Ligands: FAD, FMT, 2EH |
 |
Organism: Duganella sp. zlp-xi
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-03-29 Classification: FLAVOPROTEIN Ligands: SO4, GOL, FAD |
 |
Crystal Structure Of Sars-Cov-2 Spike Receptor-Binding Domain Bound With 6-2C Fab
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-03-01 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Crystal Structure Of The First Bromodomain Of Human Brd4 In Complex With The Inhibitor Y13157
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2022-08-10 Classification: PROTEIN BINDING Ligands: JFL, GOL |
 |
Crystal Structure Of The First Bromodomain Of Human Brd4 In Complex With The Inhibitor Y13153
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2022-08-10 Classification: PROTEIN BINDING Ligands: JFR, GOL |

