Search Count: 365
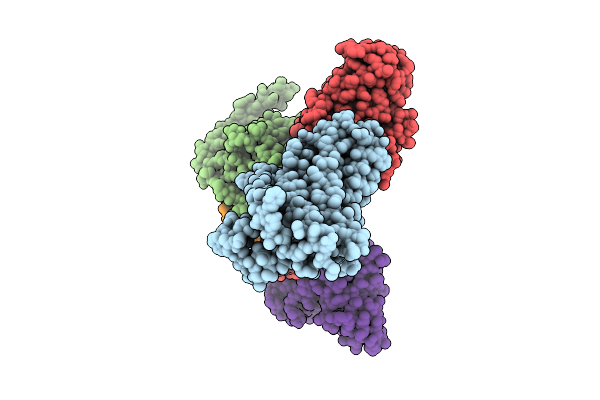 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
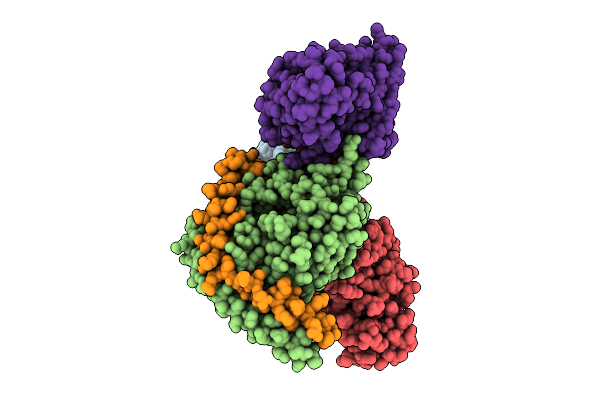 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: NIO |
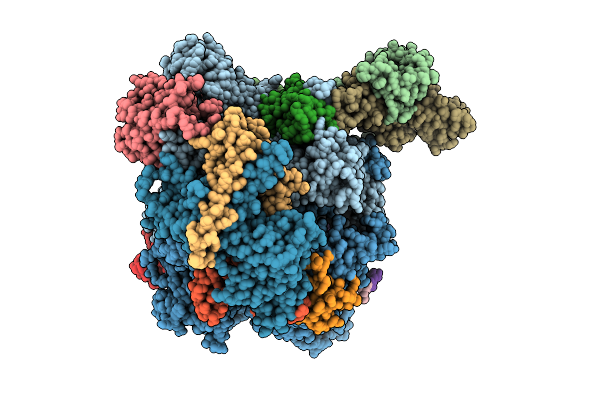 |
The Structure Of Rna Polymerase Ii Elongation Complex Paused At N-1 State By Actinomycin D.
Organism: Saccharomyces cerevisiae s288c, Synthetic construct, Streptomyces sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSCRIPTION Ligands: ZN, MG |
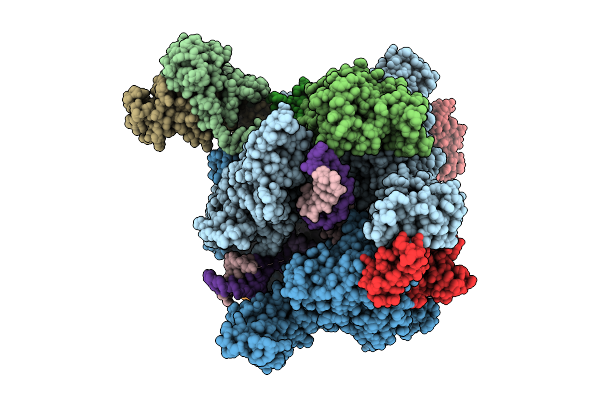 |
The Structure Of Rna Polymerase Ii Elongation Complex Paused At N-2 State By Actinomycin D.
Organism: Saccharomyces cerevisiae s288c, Synthetic construct, Streptomyces sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSCRIPTION Ligands: ZN, MG |
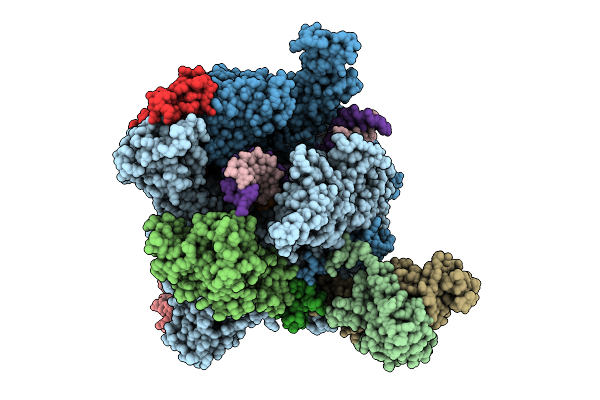 |
The Structure Of 1 Actd Bound To Rna Polymerase Ii Elongation Complex With 2 Ctg Repeats.
Organism: Synthetic construct, Saccharomyces cerevisiae s288c, Streptomyces sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSCRIPTION Ligands: ZN, MG |
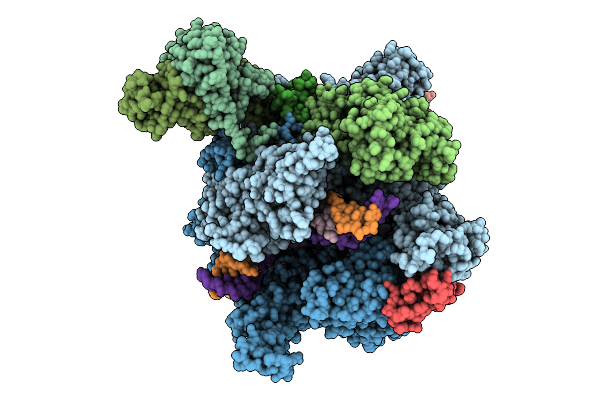 |
The Structure Of 2 Actd Bound To Rna Polymerase Ii Elongation Complex With 3 Ctg Repeats
Organism: Synthetic construct, Saccharomyces cerevisiae s288c, Streptomyces sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
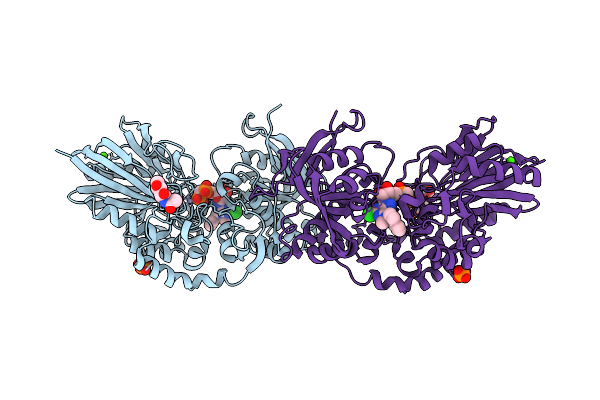
The Structure Of 3 Actd Bound To Rna Polymerase Ii Elongation Complex With 4 Ctg Repeats.
Organism: Synthetic construct, Saccharomyces cerevisiae s288c, Streptomyces sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
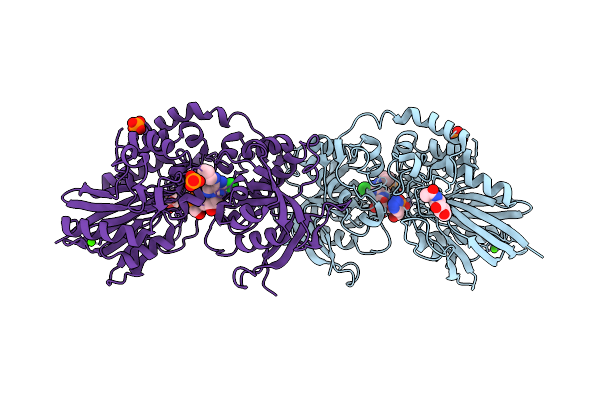
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: ZN, CA, NAG, A1JCF, PO4 |
 |
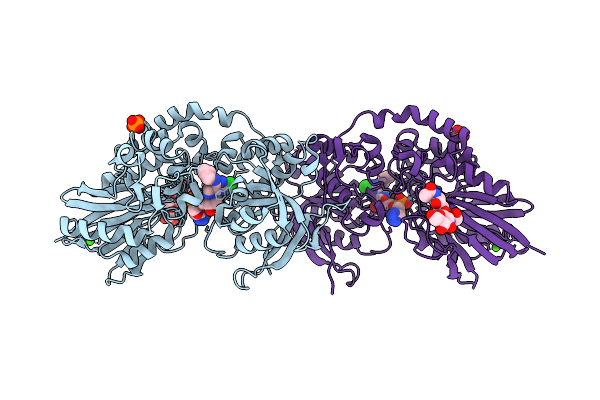
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: ZN, CA, NAG, A1JCG, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: ZN, CA, A1JCH, PO4 |
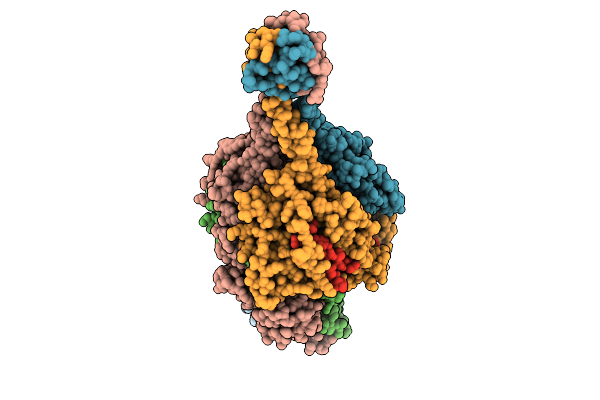 |
Respiratory Syncytial Virus Pre-F Trimer Bound By Neutralizing Antibody Pr306007
Organism: Respiratory syncytial virus a2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: STRUCTURAL PROTEIN/IMMUNE SYSTEM |
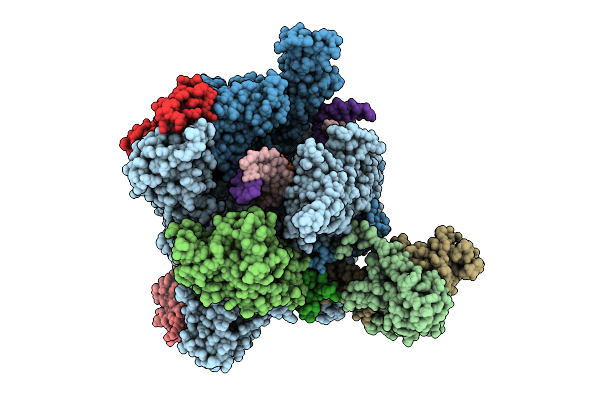 |
The Structure Of Rna Polymerase Ii Elongation Complex Paused At N-5 State By Actinomycin D.
Organism: Saccharomyces cerevisiae s288c, Synthetic construct, Streptomyces sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: ZN, MG |
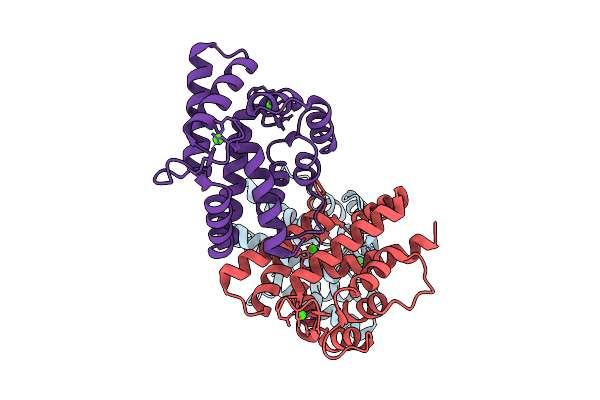 |
X-Ray Structure Of Sarcoplasmic Ca-Binding Protein (Scp), A Calcium Ion-Binding Protein From Pinctada Fucata
Organism: Pinctada fucata
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: METAL BINDING PROTEIN Ligands: CA |
 |
Organism: Yersinia pseudotuberculosis serotype o:3 (strain ypiii)
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: HYDROLASE Ligands: SRT |
 |
Organism: Synthetic construct, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Drtc) In Post-Capping State
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: VIRAL PROTEIN/RNA Ligands: ZN |
 |
Sars-Cov-2 Replication-Transcription Complex Has A Dimer Architecture (Local Drtc) In Post-Capping State
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: VIRAL PROTEIN/RNA Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: SIGNALING PROTEIN Ligands: GDP, MG, U6L, EPE |
 |
Crystal Structure Of An Agose Isomerase Mutant 5 (Tst4Ease M5) From Thermotogota Bacterium With Zn
Organism: Thermotogota bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: ISOMERASE Ligands: ZN |
 |
Crystal Structure Of A Wild-Type Tagose Isomerase (Tst4Ease Wt) From Thermotogota Bacterium Complex With Zn
Organism: Thermotogota bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: ISOMERASE Ligands: ZN |

