Search Count: 194
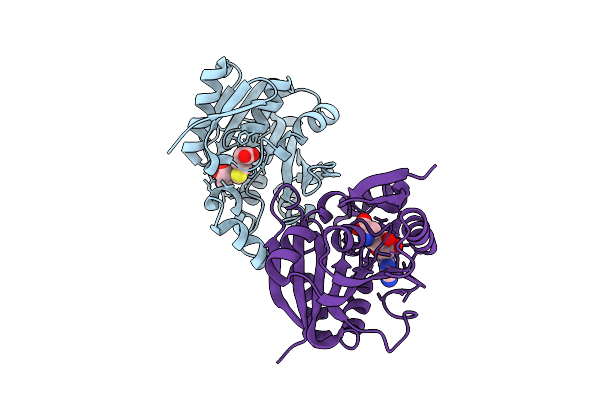 |
Crystal Structure Of The Carbamoyl N-Methyltransferase Asc-Orf2 Complexed With Sah
Organism: Amycolatopsis alba dsm 44262
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: BIOSYNTHETIC PROTEIN Ligands: SAH |
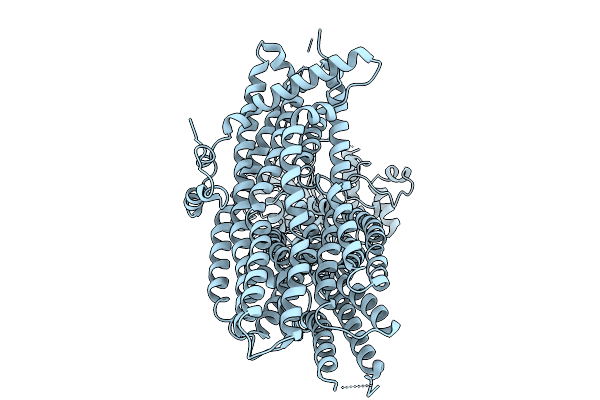 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: TRANSPORT PROTEIN |
 |
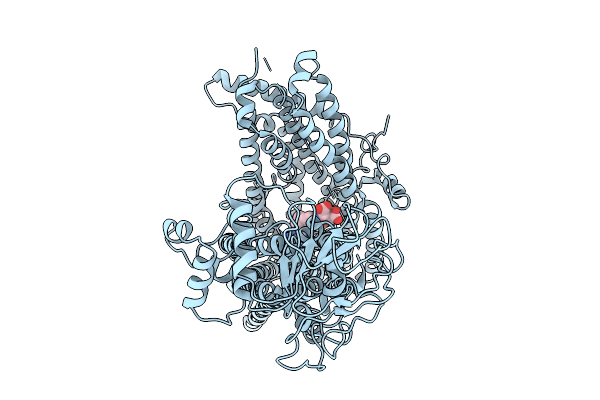
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: TRANSPORT PROTEIN Ligands: CMP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: TRANSPORT PROTEIN Ligands: MTX |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: TRANSPORT PROTEIN Ligands: P2E |
 |
Organism: Streptomyces aureocirculatus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Oxidoreductase Mutant M1 From Streptomyces Aureocirculatus
Organism: Streptomyces aureocirculatus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of An Oxidoreductase Mutant M4 From Streptomyces Aureocirculatus
Organism: Streptomyces aureocirculatus
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: OXIDOREDUCTASE |
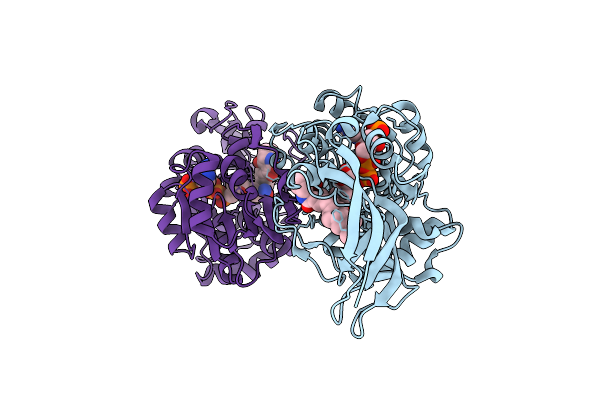 |
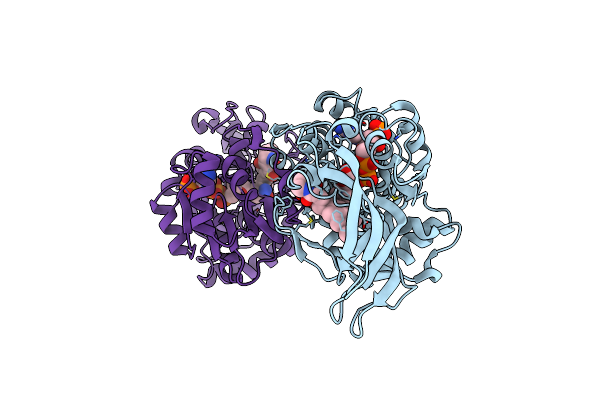
Organism: Lysobacter enzymogenes
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: BIOSYNTHETIC PROTEIN Ligands: A1EBZ, NAP |
 |
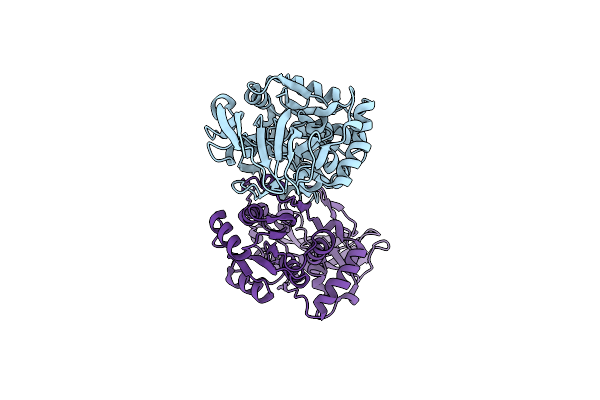
Organism: Lysobacter enzymogenes
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: BIOSYNTHETIC PROTEIN Ligands: NAP, E8T |
 |
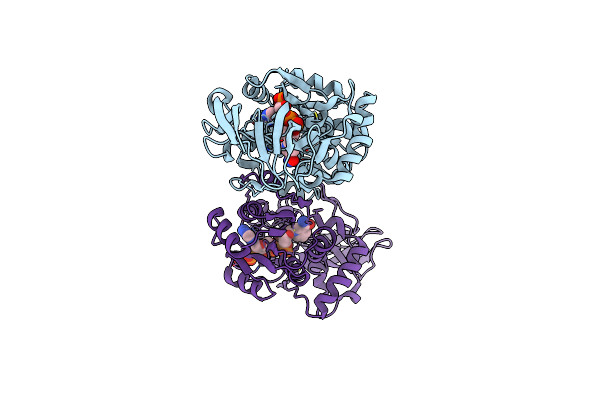
Organism: Lysobacter enzymogenes
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: BIOSYNTHETIC PROTEIN |
 |
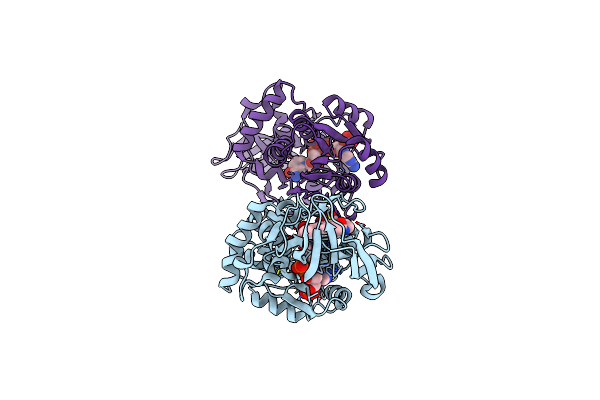
Organism: Lysobacter enzymogenes
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: BIOSYNTHETIC PROTEIN Ligands: NAP |
 |
Organism: Lysobacter enzymogenes
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: BIOSYNTHETIC PROTEIN Ligands: NAP, A1EDR |
 |
Organism: Lysobacter enzymogenes
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: BIOSYNTHETIC PROTEIN Ligands: A1EDK, NAP |
 |
Organism: Kitasatospora sp.
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4, UNL |
 |
Crystal Structure Of Ppargamma Ligand Binding Domain (Lbd) In Complex With Ncor1 Corepressor Peptide And Inverse Agonist Fx-909
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-14 Classification: NUCLEAR PROTEIN Ligands: A1CAA |
 |
Crystal Structure Of A Benzaldehyde Lyase Mutant M6 From Herbiconiux Sp. Salv-R1
Organism: Herbiconiux sp. salv-r1
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2025-02-12 Classification: LYASE Ligands: TPP, MG |
 |
Crystal Structure Of A Benzaldehyde Lyase Mutant M3 From Herbiconiux Sp. Salv-R1
Organism: Herbiconiux sp. salv-r1
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2025-02-12 Classification: LYASE Ligands: PEG, MES, TPP, MG |
 |
Organism: Sus scrofa, African swine fever virus
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: African swine fever virus, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2024-09-04 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |

