Search Count: 189
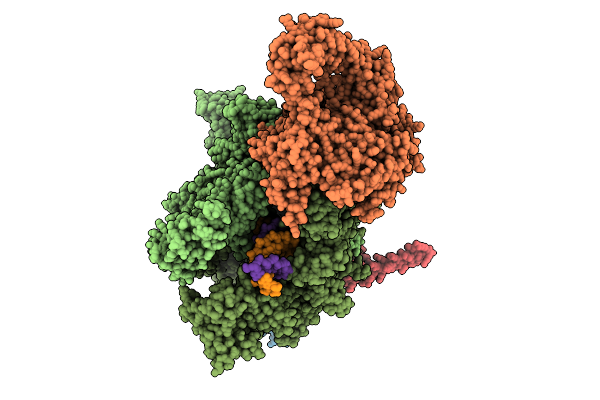 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
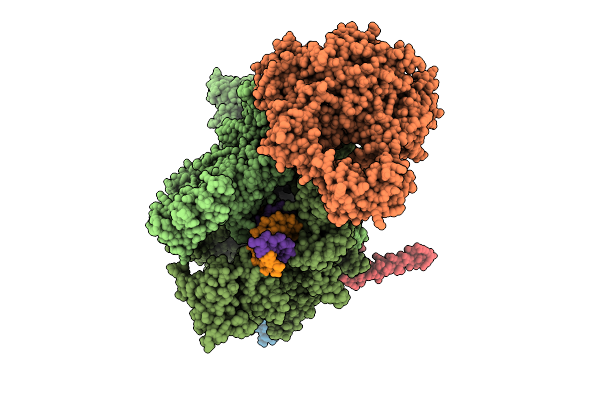 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
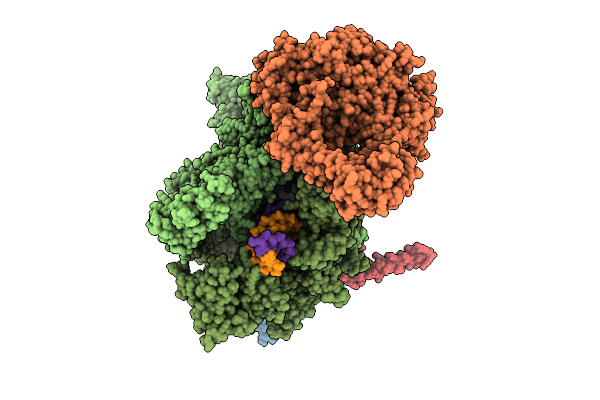 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
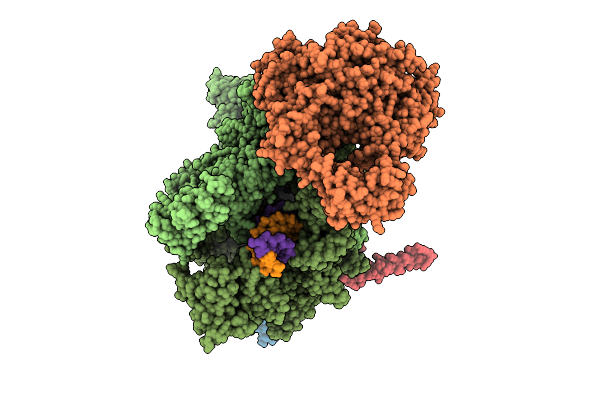 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Atp|Adp-Bound Ifasym-2 State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ATP, MG, ADP |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Adp-Bound Ifasym-3 State (Atp 37 Degrees C Treated
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ADP, MG |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Adp-Bound Ifasym-3 State (Adp 4 Degrees C Treated)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ADP, MG |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Adp-Bound Ifasym-3 (Peptidisc) State (Atp 37Degrees C Treated)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ADP |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Adp-Bound Ifasym-3 (Peptidisc) State (Adp 4Degrees C Treated)
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ADP |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Atp|Adp+Vi-Bound Occ (Vi) State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ATP, MG, ADP, VO4 |
 |
Cryo-Em Structure Of Msrv1273C/72C(E553Q) Mutant From Mycobacterium Smegmatis In The Atp-Bound Occ State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-05-14 Classification: TRANSPORT PROTEIN Ligands: ATP, MG |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Ifapo State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: TRANSPORT PROTEIN |
 |
Cryo-Em Structure Of Msrv1273C/72C From Mycobacterium Smegmatis In The Amppnp-Bound Ifasym-1 State
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: TRANSPORT PROTEIN Ligands: ANP, MG |
 |
Crystal Structure Of A Polyketide Cyclase Fasu From Streptomyces Kanamyceticus
Organism: Streptomyces kanamyceticus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2025-03-12 Classification: BIOSYNTHETIC PROTEIN Ligands: AE3, MG |
 |
Crystal Structure Of Caenorhabditis Elegans Him-8 Zf1-2-Ctd Domain In Complex With Chromosome X Pairing Center
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2024-12-11 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Crystal Structure Of Caenorhabditis Elegans Zim-2 Zf1-2-Ctd Domain In Complex With Chromosome V Pairing Center
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-12-11 Classification: DNA BINDING PROTEIN Ligands: ZN |
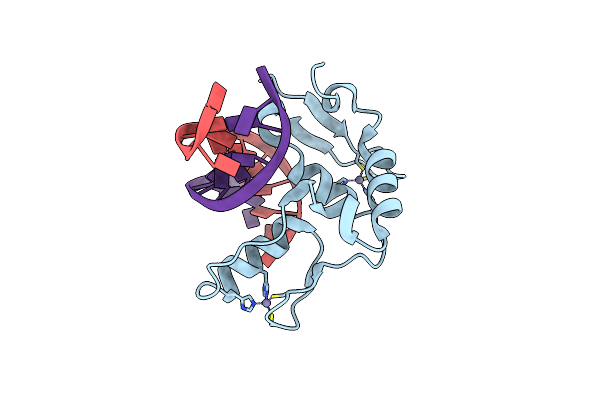 |
Crystal Structure Of Caenorhabditis Elegans Zim-1 Zf1-2-Ctd Domain In Complex With Chromosome Ii/Iii Pairing Center
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-12-11 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2024-10-23 Classification: TRANSFERASE Ligands: 6I5 |

