Search Count: 695
 |
Organism: Clostridium acetobutylicum atcc 824
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RNA |
 |
 |
Organism: Mogibacterium pumilum
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RNA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PT5, K, CA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PIO, K |
 |
Organism: Bdellovibrio bacteriovorus hd100
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: SIGNALING PROTEIN |
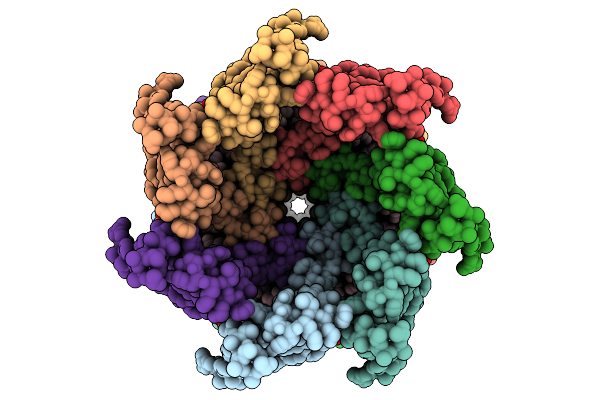 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN Ligands: PTY |
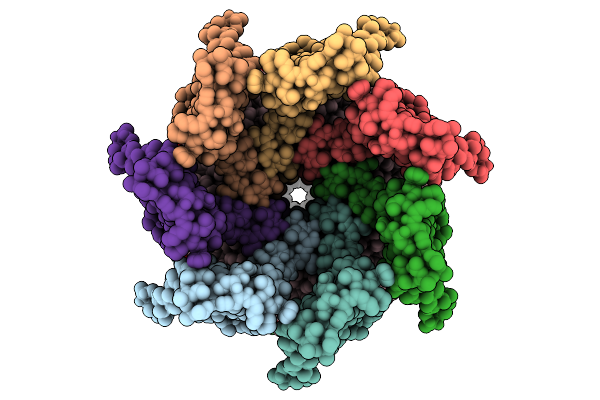 |
Cryo-Em Structure Of Mechanosensitive Channel Ynai In Dopc Nanodiscs Treated With Beta-Cyclodextrin
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN Ligands: PTY |
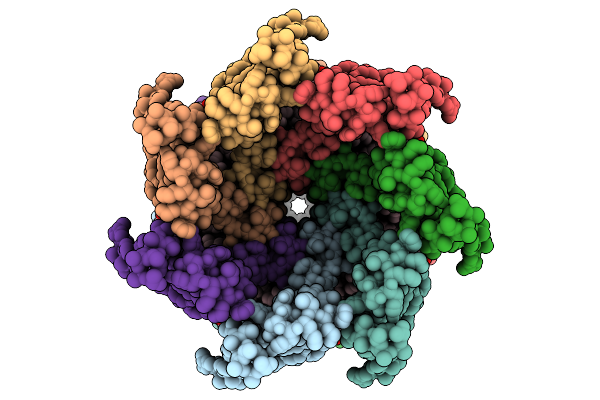 |
Cryo-Em Structure Of Mechanosensitive Channel Ynai In Dopc Nanodiscs Treated With Lysophosphatidylcholine
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN Ligands: PTY |
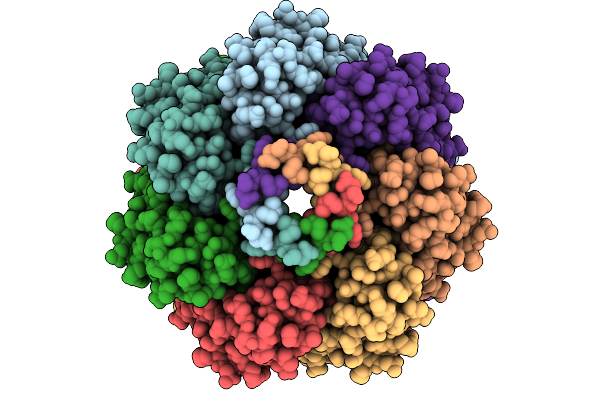 |
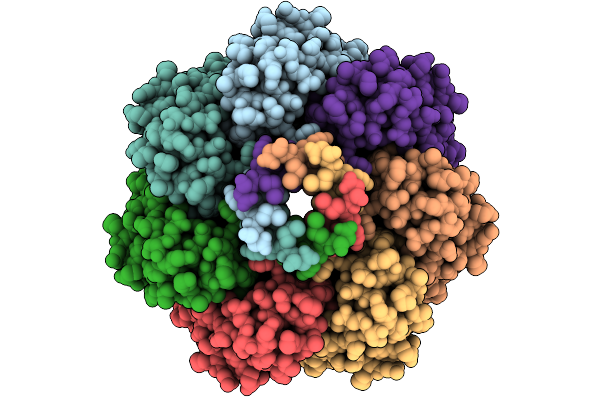
Cryo-Em Structure Of Mechanosensitive Channel Ynai A155V Mutant In Conformation 1
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN Ligands: PTY |
 |
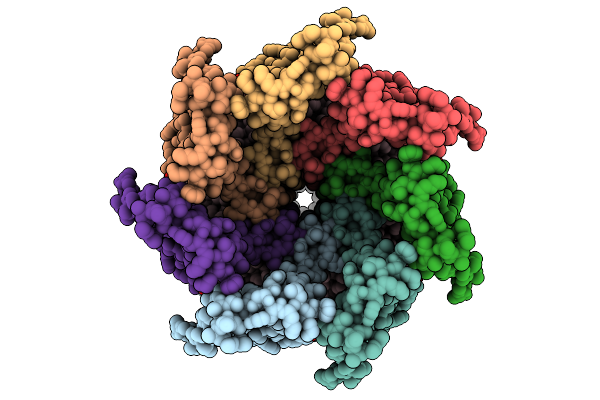
Cryo-Em Structure Of Mechanosensitive Channel Ynai A155V Mutant In Conformation 2
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN Ligands: PTY |
 |
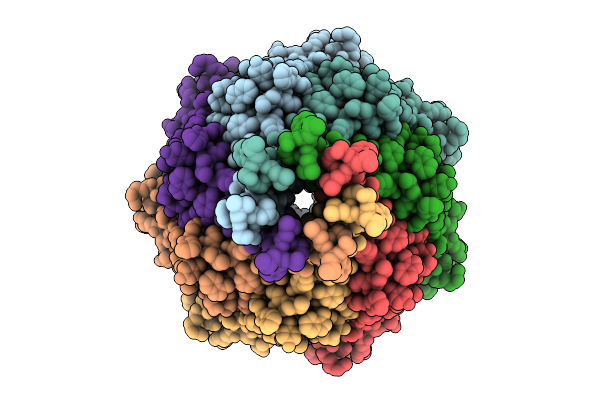
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN Ligands: PTY |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN |
 |
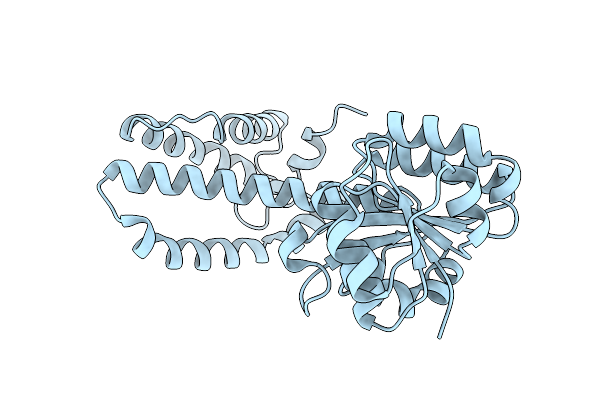
Organism: Micromonospora echinaurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
Organism: Micromonospora echinaurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE |
 |
Organism: Kutzneria albida dsm 43870
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: NDP, NA |
 |
Organism: Kutzneria albida dsm 43870
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: NDP, SO4, PE8, NA |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Zea Mays 3-Phosphoglycerate Dehydrogenase S282L Mutant
Organism: Zea mays
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: OXIDOREDUCTASE |
 |
Organism: Shewanella oneidensis mr-1
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: STRUCTURAL PROTEIN |

