Maintenance Mode: Some services may turn out to be unavailable. We apologize for the inconvenience!
Maintenance Mode: Some services may turn out to be unavailable. We apologize for the inconvenience!
 |
Sivtal Integrase In Complex With Rna Stem-Loop (Focused Refinement Of The Filament Repeat Unit)
Organism: Simian immunodeficiency virus, Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Cryoem Reconstruction Of Integrase Filament At The Lumen Of Native Hiv-1 Cores (Box Size 34.2 Nm)
Organism: Human immunodeficiency virus 1
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN Ligands: ZN, IHP |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: HYDROLASE Ligands: 4BW |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: HYDROLASE |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: HYDROLASE |
 |
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: FLAVOPROTEIN Ligands: FMN |
 |
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: FLAVOPROTEIN Ligands: FAD |
 |
Organism: Acinetobacter baumannii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: HYDROLASE Ligands: 4BW |
 |
Organism: Xenopus laevis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: DNA BINDING PROTEIN/DNA Ligands: ZN |
 |
Organism: Clostridium acetobutylicum atcc 824
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RNA |
 |
 |
Organism: Mogibacterium pumilum
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RNA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PT5, K, CA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PIO, K |
 |
Organism: Bdellovibrio bacteriovorus hd100
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: SIGNALING PROTEIN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN Ligands: PTY |
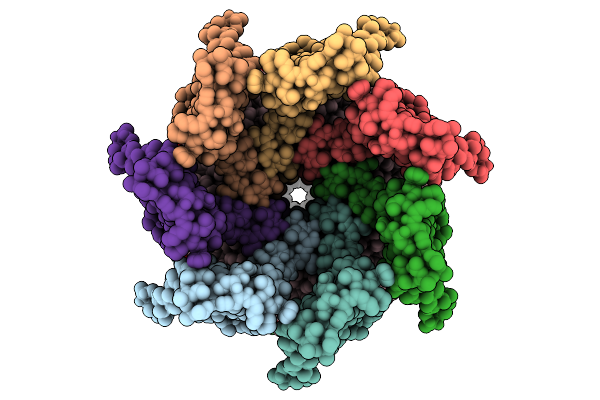 |
Cryo-Em Structure Of Mechanosensitive Channel Ynai In Dopc Nanodiscs Treated With Beta-Cyclodextrin
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN Ligands: PTY |
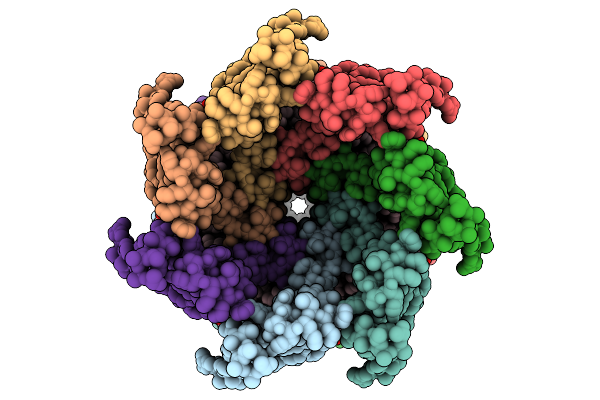 |
Cryo-Em Structure Of Mechanosensitive Channel Ynai In Dopc Nanodiscs Treated With Lysophosphatidylcholine
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN Ligands: PTY |
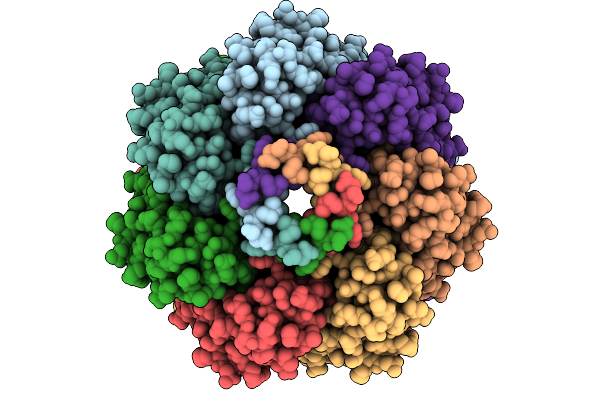 |
Cryo-Em Structure Of Mechanosensitive Channel Ynai A155V Mutant In Conformation 1
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN Ligands: PTY |
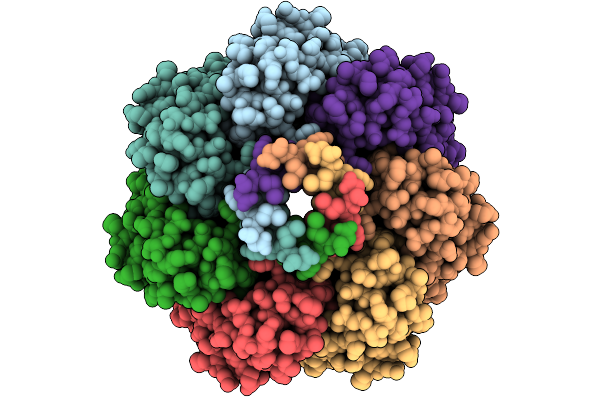 |
Cryo-Em Structure Of Mechanosensitive Channel Ynai A155V Mutant In Conformation 2
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-08-27 Classification: MEMBRANE PROTEIN Ligands: PTY |