Search Count: 16
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: MEMBRANE PROTEIN |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:3.90 Å Release Date: 2019-04-03 Classification: PLANT PROTEIN Ligands: U5P |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:4.25 Å Release Date: 2019-04-03 Classification: PLANT PROTEIN Ligands: U5P |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2019-04-03 Classification: PLANT PROTEIN Ligands: ADP |
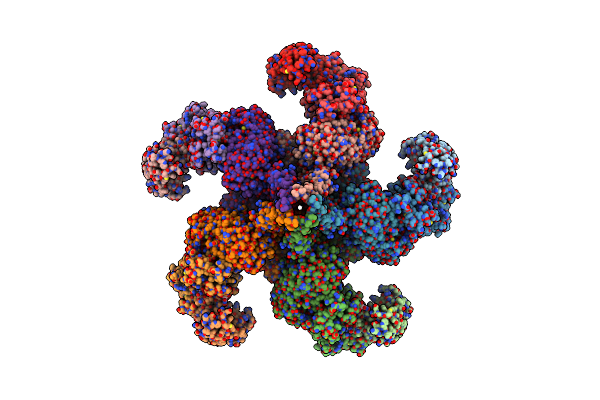 |
Reconstitution And Structure Of A Plant Nlr Resistosome Conferring Immunity
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2019-03-20 Classification: PLANT PROTEIN Ligands: U5P, DTP |
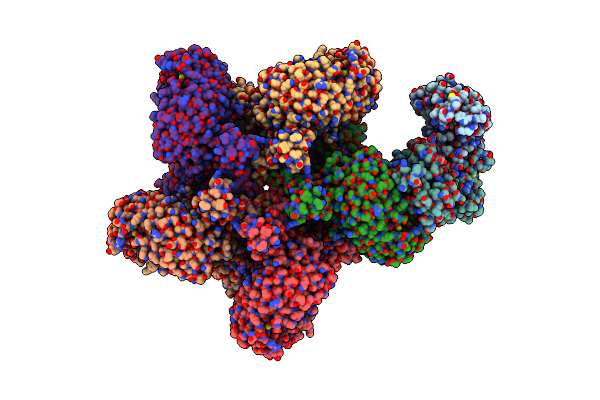 |
Reconstitution And Structure Of A Plant Nlr Resistosome Conferring Immunity
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:3.70 Å Release Date: 2019-03-20 Classification: PLANT PROTEIN Ligands: U5P, DTP |
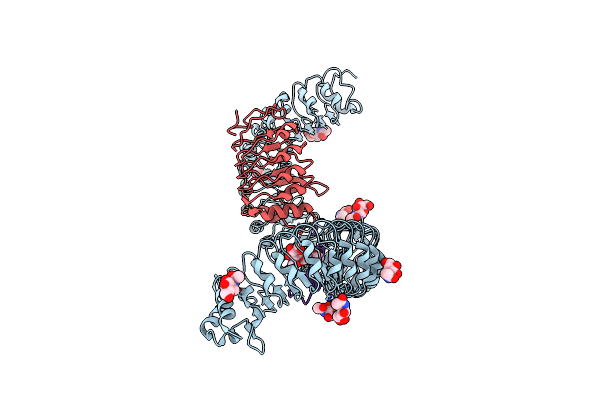 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2013-12-04 Classification: TRANSFERASE/TRANSFERASE RECEPTOR Ligands: NAG, SO4 |
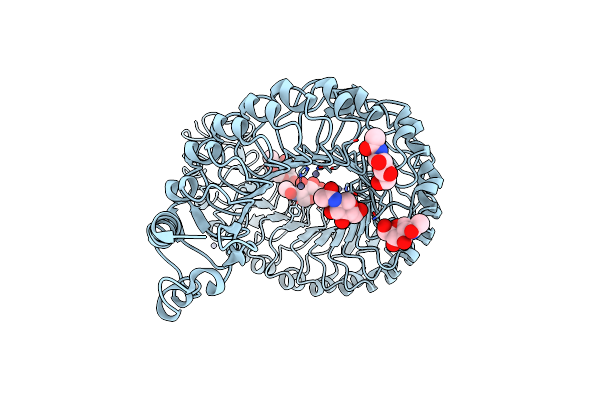 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:4.00 Å Release Date: 2013-12-04 Classification: TRANSFERASE Ligands: NAG, ZN |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2012-06-27 Classification: TRANSFERASE Ligands: NAG |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2012-06-27 Classification: TRANSFERASE |
 |
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-12-18 Classification: LYASE |
 |
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-12-18 Classification: LYASE Ligands: SO4 |
 |
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-12-18 Classification: LYASE Ligands: TLA |
 |
Organism: Salmonella typhimurium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2007-12-18 Classification: LYASE |
 |
Structural Basis For Activation Of Plant Immunity By Bacterial Effector Protein Avrpto
Organism: Pseudomonas syringae, Solanum pimpinellifolium
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2007-08-21 Classification: TRANSFERASE |
 |
Structure Of Holo-Glyceraldehyde-3-Phosphate-Dehydrogenase From Palinurus Versicolor Refined 2.0 Angstrom Resolution
Organism: Palinurus versicolor
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1998-09-16 Classification: OXIDOREDUCTASE Ligands: SO4, NAD |

