Search Count: 935
 |
Organism: Sordaria araneosa
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: BIOSYNTHETIC PROTEIN Ligands: A1LYT, MPD |
 |
The Structure Of Sdng Covalently Binding With The Cope Rearrangement Product
Organism: Sordaria araneosa
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: BIOSYNTHETIC PROTEIN Ligands: MPD, A1LZG |
 |
Organism: Sordaria araneosa
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: BIOSYNTHETIC PROTEIN Ligands: A1LZH, A1LZK |
 |
Organism: Sordaria araneosa
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: BIOSYNTHETIC PROTEIN Ligands: MPD |
 |
Organism: Sordaria araneosa
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: BIOSYNTHETIC PROTEIN Ligands: A1LYT |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: ANTITUMOR PROTEIN Ligands: ZN |
 |
Structural Basis For The Polymer-Protein Binding Mechanism Of Polyvinyl Alcohol Esterase
Organism: Comamonas sp. nyz500
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: SO4, ACT, GOL, CL, NA |
 |
Structural Basis For The Polymer-Protein Binding Mechanism Of Polyvinyl Alcohol Esterase
Organism: Comamonas sp. nyz500
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: GOL |
 |
Structural Basis For The Polymer-Protein Binding Mechanism Of Polyvinyl Alcohol Esterase
Organism: Comamonas sp. nyz500
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: CL, GOL, ACT |
 |
Structural Basis For The Polymer-Protein Binding Mechanism Of Polyvinyl Alcohol Esterase
Organism: Comamonas sp. nyz500
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: CL, A1EGH, ACT, PO4 |
 |
Structural Basis For The Polymer-Protein Binding Mechanism Of Polyvinyl Alcohol Esterase
Organism: Comamonas sp. nyz500
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: HYDROLASE Ligands: A1EGL, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: IMMUNE SYSTEM |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: LYASE Ligands: FES |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: TRANSFERASE Ligands: A1L5T, SO4, CO |
 |
Apg Mutant Enzyme D448N Of Acarbose Hydrolase From Human Gut Flora K. Grimontii Td1,Complex With Acarbose
Organism: Klebsiella grimontii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: HYDROLASE |
 |
Apg Mutant Enzyme D448N Of The Human Gut Flora K. Grimontii Td1 Acarbose Hydrolase
Organism: Klebsiella grimontii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: HYDROLASE |
 |
Structure Of A Truncated Loopa Mutant From The Human Gut Flora K. Grimontii Apg In Complex With Glucose
Organism: Klebsiella grimontii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: HYDROLASE Ligands: GLC |
 |
Structure Of A Truncated Loopb Mutant From The Human Gut Flora K. Grimontii Apg
Organism: Klebsiella grimontii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: HYDROLASE |
 |
Structure Of Truncated Loopa And Loopb Mutants From The Human Gut Flora K. Grimontii Apg
Organism: Klebsiella grimontii
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: HYDROLASE |
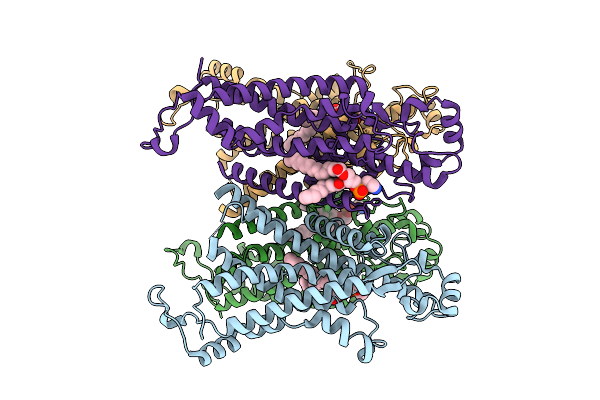 |
Organism: Medicago truncatula
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: MEMBRANE PROTEIN Ligands: PTY |

