Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Transcription Factor Deltafosb/Jund Bzip Domain In Complex With An Effector Molecule
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: CELL ADHESION Ligands: A1CAO, CL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/HYDROLASE |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/HYDROLASE |
 |
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: LYASE Ligands: PO4, CL |
 |
Gamma-Lyase Cndf In Complex With Pyridoxal 5'-Phosphate, L-Homoserine And Ethyl Acetoacetate
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: LYASE Ligands: HSE, EAC |
 |
Organism: Penicillium citrinum
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: LYASE Ligands: PO4, EAC |
 |
 |
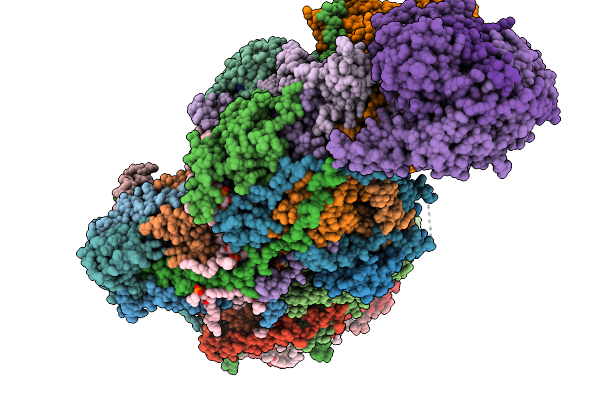 |
Complex I From Respirasome Closed State 1 Bound By Metformin And Coq10, Alternative Orientation (Sc-Metc1-Ii)
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, PC1, NDP, PEE, ZMP, ADP, PLX, 3PE, PX2, U10, SF4, FES, MG, MF8, ZN, FMN |
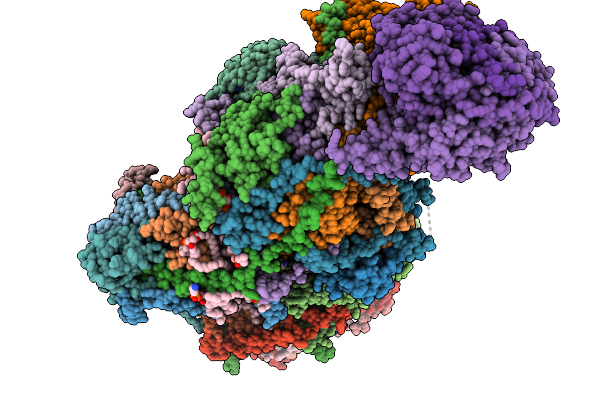 |
Complex I From Respirasome Closed State 1 Bound By Metformin And Coq10 (Sc-Metc1-I)
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, PC1, NDP, PEE, ZMP, ADP, PLX, 3PE, U10, SF4, FES, MG, MF8, ZN, FMN |
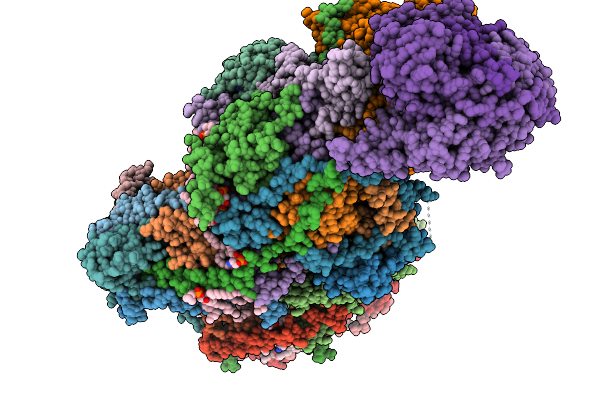 |
Complex I From Respirasome Closed State 1 Bound By Metformin (Sc-Metc1-Iii)
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, PC1, NDP, PEE, ZMP, ADP, PLX, 3PE, SF4, FES, MG, MF8, ZN, FMN |
 |
 |
 |
 |
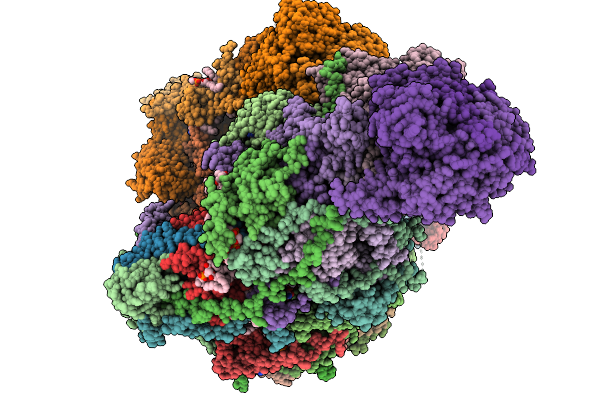
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, NDP, PEE, ZMP, ADP, PLX, 3PE, XEW, 6F6, SF4, FES, MG, ZN, FMN |
 |
Complex I From Respirasome Closed State 1 Bound By Metformin And Coq10 (Sc-Metc1-Iv)
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, NDP, PEE, PC1, ZMP, ADP, PLX, 3PE, U10, SF4, FES, MG, MF8, ZN, FMN |
 |
Complex I From Respirasome Closed State 1 Bound By Metformin And Coq10 (Sc-Metc1-V)
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: CDL, NDP, PEE, ZMP, ADP, PLX, 3PE, PC1, SF4, FES, MG, ZN, U10, MF8, FMN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: A1IZR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: MG, A1IZW, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: HYDROLASE Ligands: A1IZX |