Search Count: 482
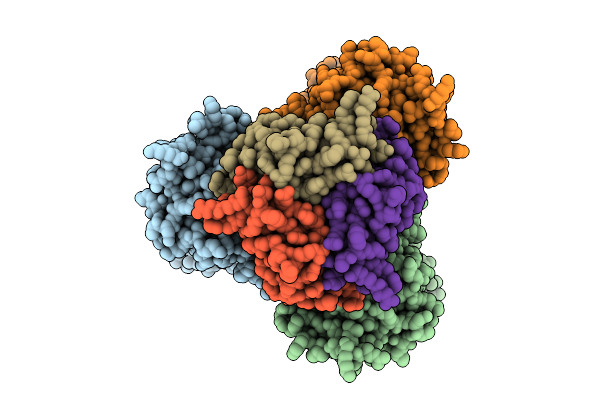 |
Cryo-Em Structure Of Bicarbonate Transporter Sbta In Complex With Pii-Like Signaling Protein Sbtb (T-Loop Truncation) From Synechocystis Sp. Pcc 6803
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: TRANSPORT PROTEIN Ligands: BCT |
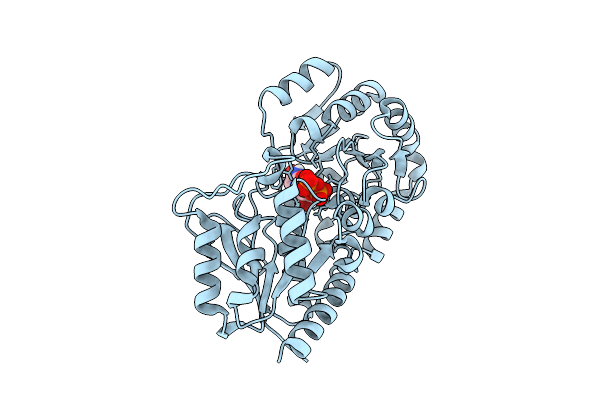 |
Udp-Binding Psbglut, A Tetrahydrobiopterin Glucosyltransferase From Pseudanabaena Sp. Chao 1811
Organism: Pseudanabaena sp. chao 1811
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: UDP |
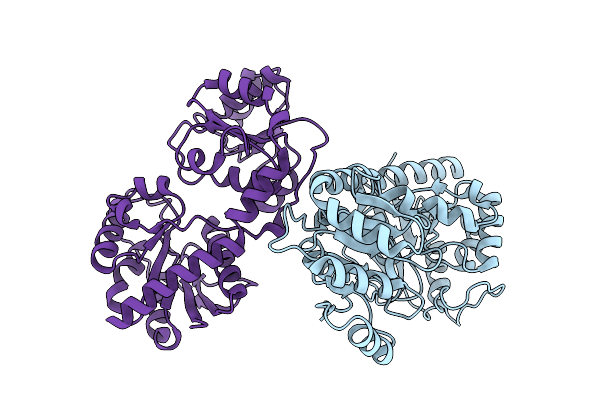 |
Udp-Glc:Tetrahydrobiopterin Glucosyltransferase From Pseudanabaena Sp. Chao 1811
Organism: Pseudanabaena sp. chao 1811
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE |
 |
Organism: Pseudanabaena phage pan3
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRUS |
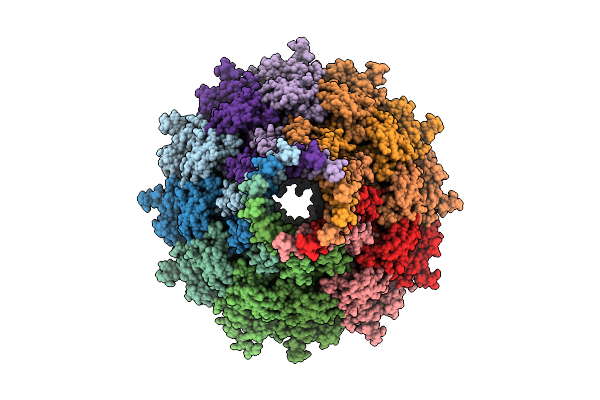 |
Organism: Pseudanabaena phage pan3
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRAL PROTEIN |
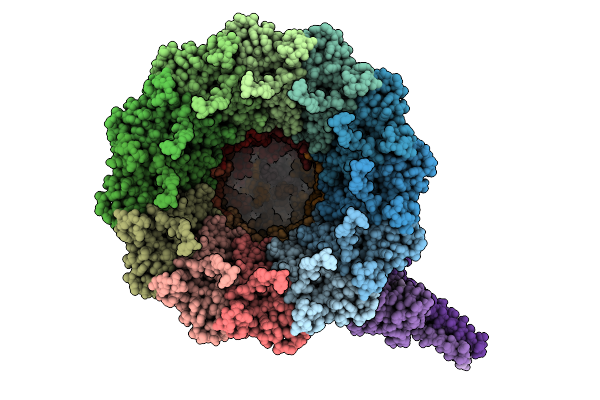 |
Organism: Pseudanabaena phage pan3
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRAL PROTEIN |
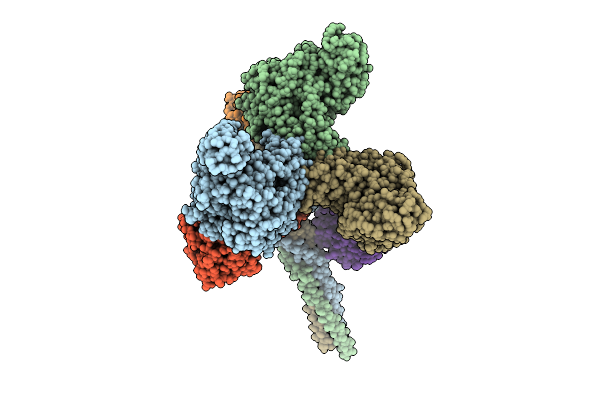 |
Organism: Pseudanabaena phage pan3
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRAL PROTEIN |
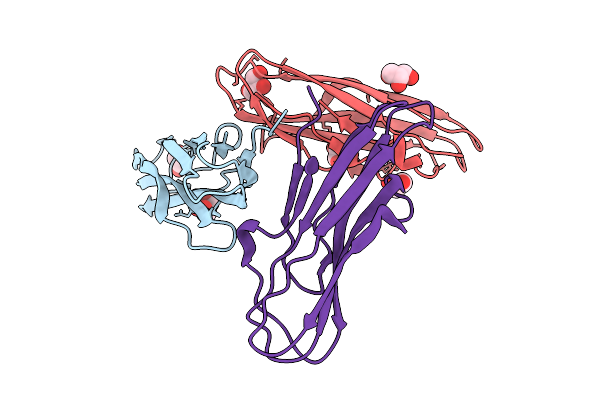 |
Organism: Pseudanabaena phage pan3
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRUS Ligands: IOD, GOL |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: RIBOSOME |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: DE NOVO PROTEIN Ligands: IOD |
 |
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: DE NOVO PROTEIN Ligands: LMT |
 |
Cryo-Em Structure Of A De Novo Designed Voltage-Gated Anion Channel (Dvgac)
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: DE NOVO PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-08-13 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: DNA BINDING PROTEIN/DNA/RNA Ligands: MG |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: DNA BINDING PROTEIN/DNA/RNA |
 |
Crystal Structure Of Human Glutaminyl Cyclase In Complex With N-(2-(1H-Imidazol-5-Yl)Ethyl)-4-Methoxybenzenesulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: TRANSFERASE Ligands: ZN, A1EFY |
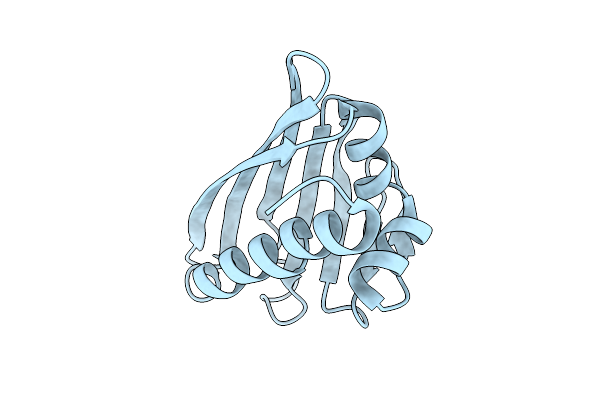 |
Benzbromarone Interferes With The Interaction Between Hsp90 And Aha1 By Interacting With Both Of Them
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: STRUCTURAL PROTEIN |
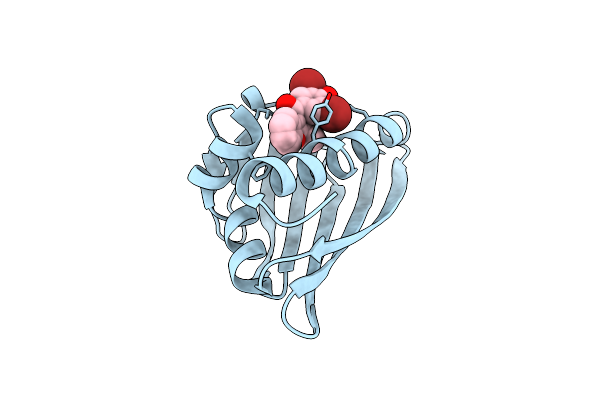 |
Benzbromarone Interferes With The Interaction Between Hsp90 And Aha1 By Interacting With Both Of Them
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: STRUCTURAL PROTEIN Ligands: R75 |
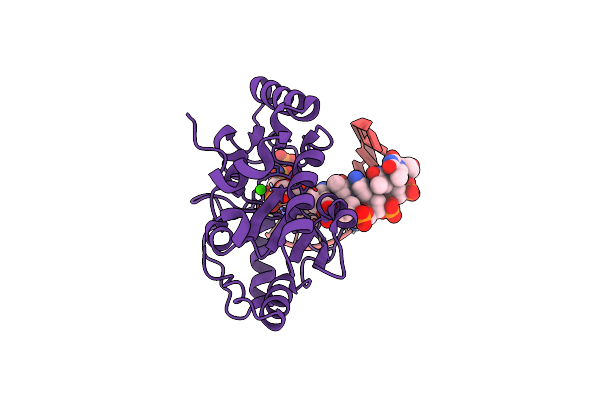 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2025-04-16 Classification: DNA BINDING PROTEIN/DNA Ligands: CA |
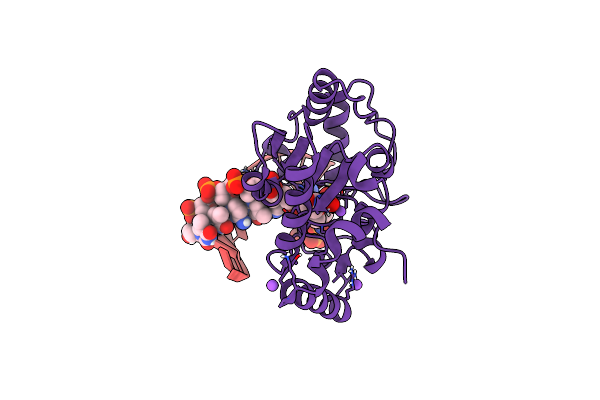 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2025-04-16 Classification: DNA BINDING PROTEIN/DNA |

