Search Count: 31
 |
Organism: Deinococcus geothermalis
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2024-03-13 Classification: HYDROLASE Ligands: SO4, MN |
 |
Organism: Deinococcus geothermalis dsm 11300, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-03-13 Classification: HYDROLASE/DNA Ligands: MN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.50 Å Release Date: 2023-12-06 Classification: TRANSFERASE Ligands: ZN, GDP, WYL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2023-12-06 Classification: TRANSFERASE Ligands: GDP, MG, WYU |
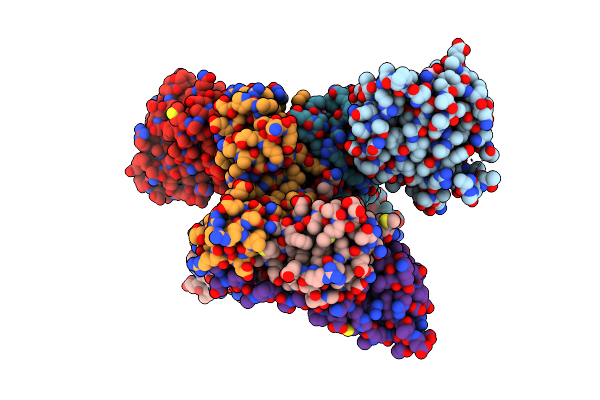 |
Crystal Structure Of Ligand Acbi3 In Complex With Kras G12D C118S Gdp And Pvhl:Elonginc:Elonginb Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2023-12-06 Classification: GENE REGULATION Ligands: PO4, WYL, GDP, MG |
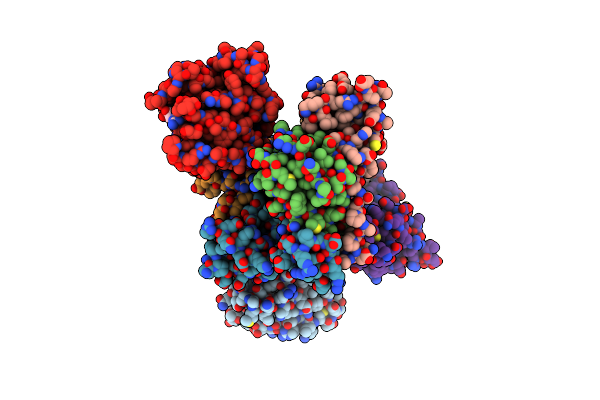 |
Crystal Structure Of Compound 3 In Complex With Kras G12V C118S Gdp And Pvhl:Elonginc:Elonginb
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-12-06 Classification: ONCOPROTEIN Ligands: GDP, X4R, MG |
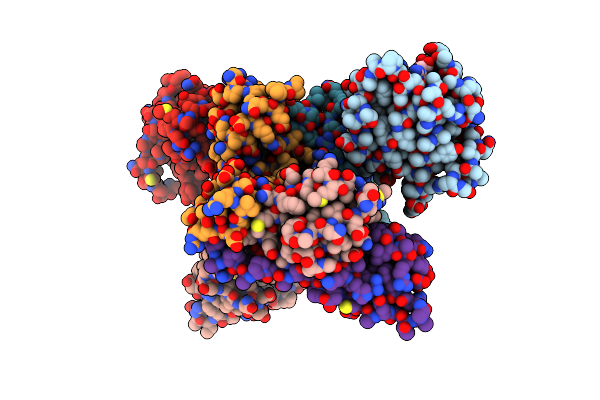 |
Crystal Structure Of Compound 4 In Complex With Kras G12V C118S Gdp And Pvhl:Elonginc:Elonginb
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2023-12-06 Classification: ONCOPROTEIN Ligands: X53, GDP, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2021-11-24 Classification: ANTITUMOR PROTEIN Ligands: GOL, NAG |
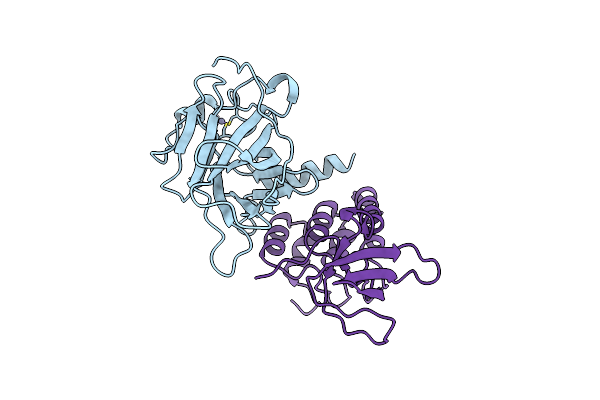 |
Crystal Structure Of A Complex Between The Dna-Binding Domain Of P53 And The Carboxyl-Terminal Conserved Region Of Iaspp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.23 Å Release Date: 2019-10-30 Classification: ANTITUMOR PROTEIN Ligands: ZN |
 |
Organism: Pseudoalteromonas sp. cf6-2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-10-16 Classification: HYDROLASE Ligands: ZN, GOL |
 |
Organism: Mus musculus, Felis catus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2018-04-04 Classification: IMMUNE SYSTEM Ligands: CA |
 |
Crystal Structure Of Trypanosoma Brucei Adometdc/Prozyme Heterodimer In Complex With Inhibitor Cgp 40215
Organism: Trypanosoma brucei brucei (strain 927/4 gutat10.1)
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2017-01-11 Classification: LYASE Ligands: B3P, CGQ, PUT |
 |
Organism: Trypanosoma brucei brucei (strain 927/4 gutat10.1)
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2017-01-11 Classification: LYASE Ligands: B3P, PUT |
 |
Organism: Trypanosoma brucei brucei
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2016-12-28 Classification: LYASE |
 |
Hepatitis B Virus Core Protein Y132A Mutant In Complex With 4-Methyl Heteroaryldihydropyrimidine
Organism: Hepatitis b virus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-08-10 Classification: VIRAL PROTEIN Ligands: IPA, 6XU, GOL, CL |
 |
Organism: Enterovirus a71
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2016-03-30 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 5GH |
 |
Organism: Enterovirus a71
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2016-03-30 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 5E8 |
 |
Organism: Enterovirus a71
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2016-03-30 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 5E9 |
 |
Organism: Enterovirus a71
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2016-03-30 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 5EB |
 |
Organism: Enterovirus a71
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2016-03-30 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 5EC |

