Search Count: 39
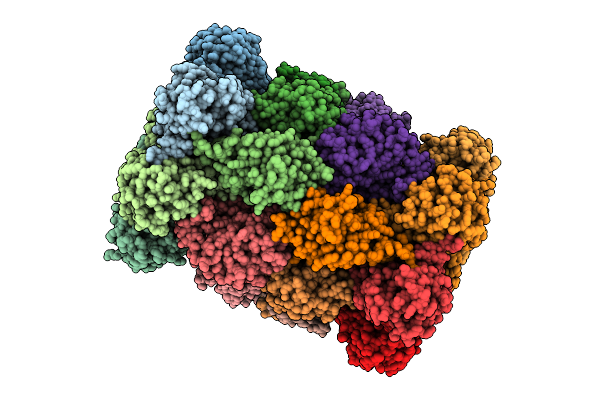 |
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: CYTOSOLIC PROTEIN Ligands: A1B74 |
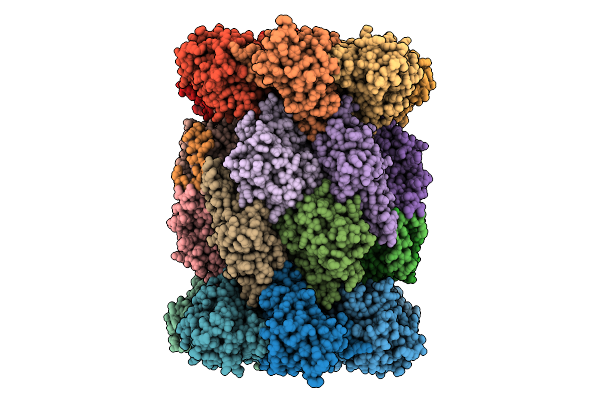 |
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: CYTOSOLIC PROTEIN Ligands: A1B73 |
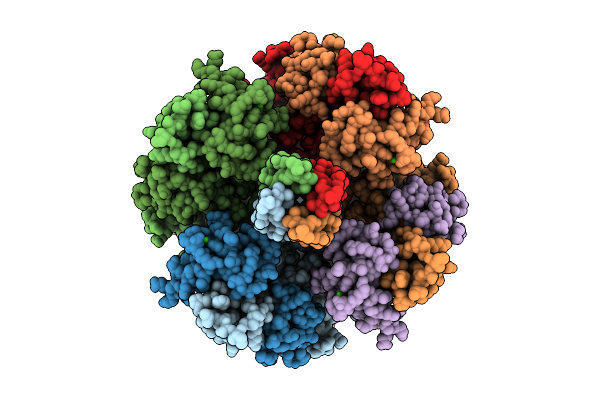 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PT5, K, CA |
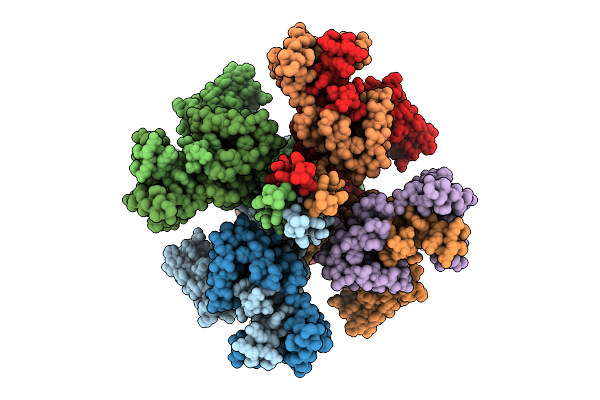 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PIO, K |
 |
Structure Of Epstein-Barr Virus Major Glycoprotein Gp350 In Complex With The Receptor Cr2
Organism: Homo sapiens, Human gammaherpesvirus 4
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: VIRAL PROTEIN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of Ebv Ghgl-Gp42 In Complex With Mabs 3E8 And 5E3 (Localized Refinement)
Organism: Human gammaherpesvirus 4, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of Ebv Ghgl-Gp42 In Complex With Mab 10E4 (Localized Refinement)
Organism: Human gammaherpesvirus 4, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of Ebv Ghgl-Gp42 In Complex With Mab 6H2 (Localized Refinement)
Organism: Human gammaherpesvirus 4, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of A M61 Aminopeptidase Family Member From Myxococcus Fulvus
Organism: Myxococcus fulvus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-06-07 Classification: HYDROLASE Ligands: PG4, P6G, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-12-14 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-12-14 Classification: MEMBRANE PROTEIN Ligands: K, I0S |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-12-14 Classification: MEMBRANE PROTEIN Ligands: K, I0S, PIO |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-12-14 Classification: MEMBRANE PROTEIN Ligands: K, I0S, PIO |
 |
Organism: Mangifera indica
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: UPG |
 |
Organism: Mangifera indica
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: UDP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2022-07-06 Classification: TRANSFERASE Ligands: 5ZH |
 |
Organism: Ralstonia solanacearum (strain gmi1000)
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2021-11-17 Classification: TRANSFERASE |

