Search Count: 608
 |
Organism: Rotavirus
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: VIRAL PROTEIN |
 |
Organism: Rotavirus
Method: X-RAY DIFFRACTION Release Date: 2025-07-09 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of Meso-Diaminopimelate Dehydrogenase From Bacillus Thermozeamaize Mutant M9 Complexed With Nadp+
Organism: Bacillus thermozeamaize
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2025-05-21 Classification: OXIDOREDUCTASE Ligands: NAP, OXM, IMD, ACY |
 |
Organism: Xanthomonas phage phixacjx1
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: VIRUS |
 |
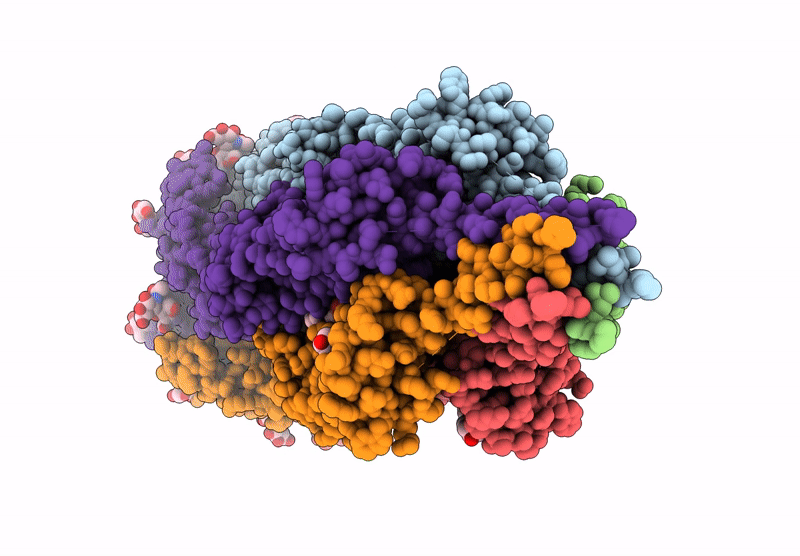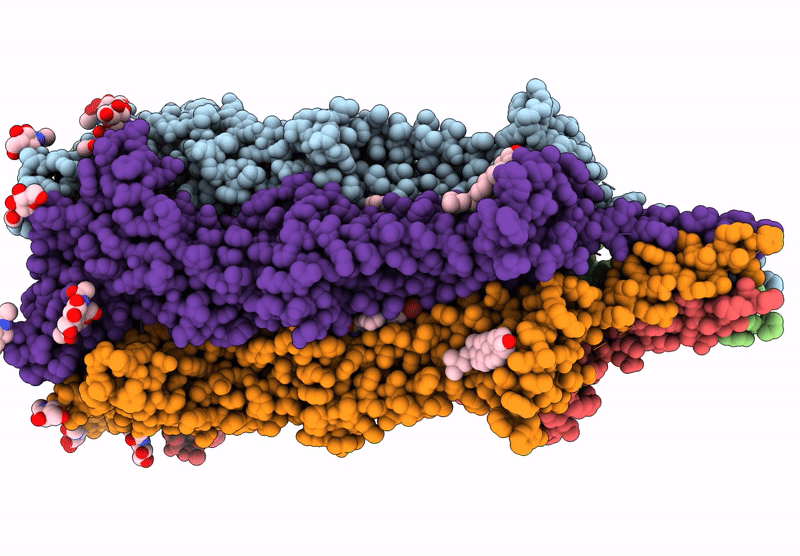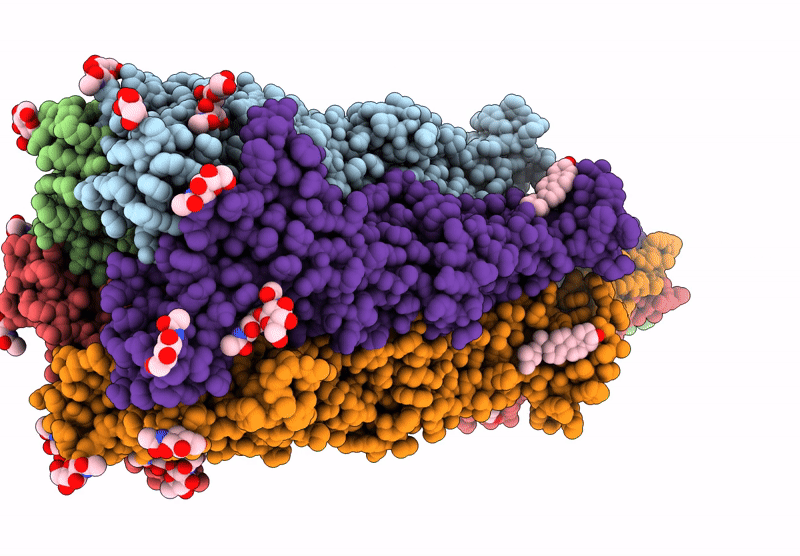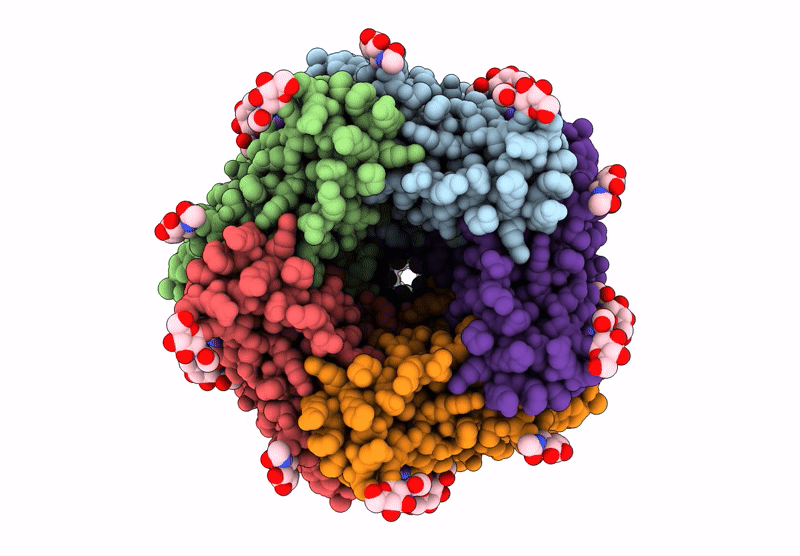
The Composite Cryo-Em Structure Of The Head-To-Tail Connector And Head-Proximal Tail Components Of Bacteriophage Phixacjx1
Organism: Xanthomonas phage phixacjx1
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: VIRUS |
 |
Human Alpha 7 Nicotinic Acetylcholine Receptor In Complex With Gat107 And Calcium (Desensitized State)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: YLI, CA, NAG, CLR |
 |
Human Alpha 7 Nicotinic Acetylcholine Receptor In Complex With Gat107 And Calcium (Open State)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: NAG, CA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN Ligands: PIO |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN Ligands: 9MF, PIO |
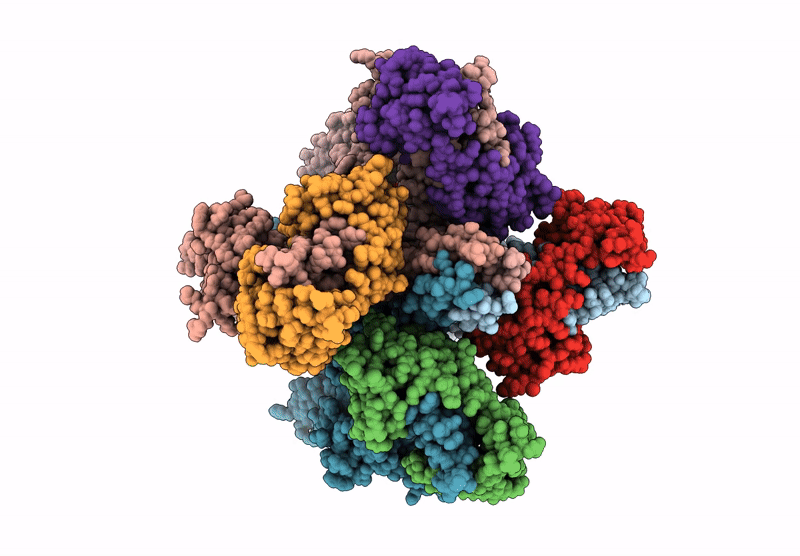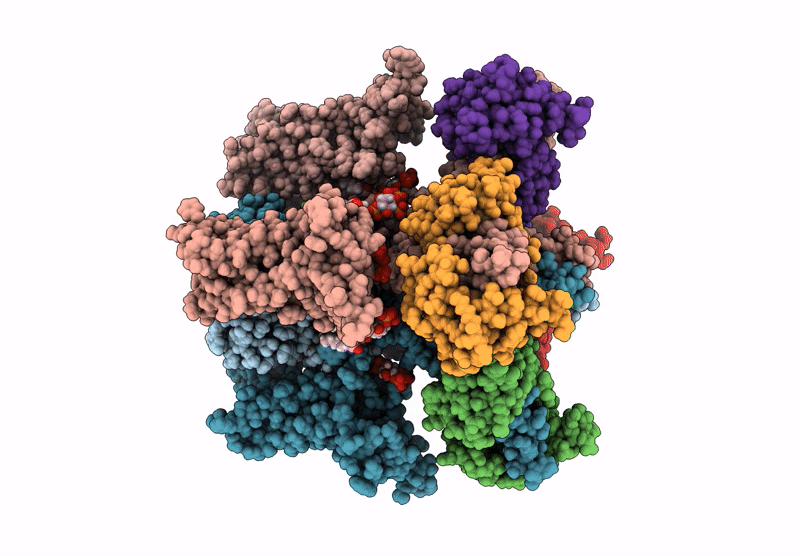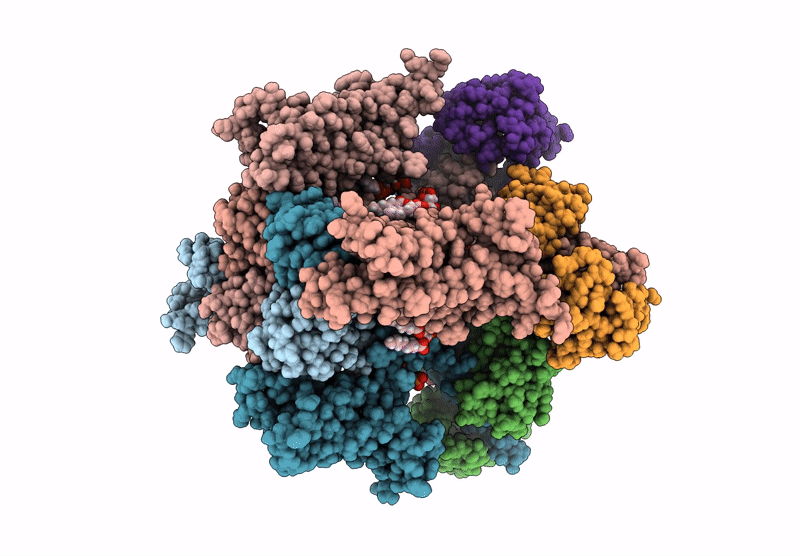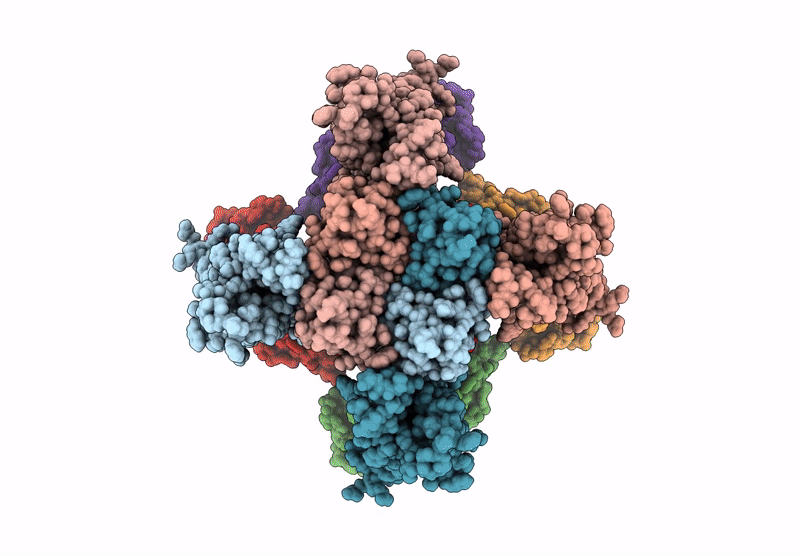 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN Ligands: PIO, 9MF |
 |
Organism: Lysinibacillus composti
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2025-04-09 Classification: OXIDOREDUCTASE Ligands: EDO, NAD |
 |
Crystal Structure Of Enl Yeats In Complex With Histone H3 Methacrylated At K18
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: TRANSCRIPTION Ligands: MES |
 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2025-04-02 Classification: OXIDOREDUCTASE Ligands: FAD, NDP |
 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: FAD, NDP |
 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: NDP, FAD |
 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.63 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: FAD, NAD |
 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2025-03-26 Classification: OXIDOREDUCTASE Ligands: NAI, FAD |
 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: FAD, NAI |
 |
Structure Of Cyclohexanone Monooxygenase Mutant From Acinetobacter Calcoaceticus
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: FAD, NDP |

