Search Count: 451
 |
The 1:1 Cryo-Em Structure Of Bap1/Asxl1-K351Ub In Complex With H2Ak119Ub Nucleosome
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: NUCLEAR PROTEIN/DNA |
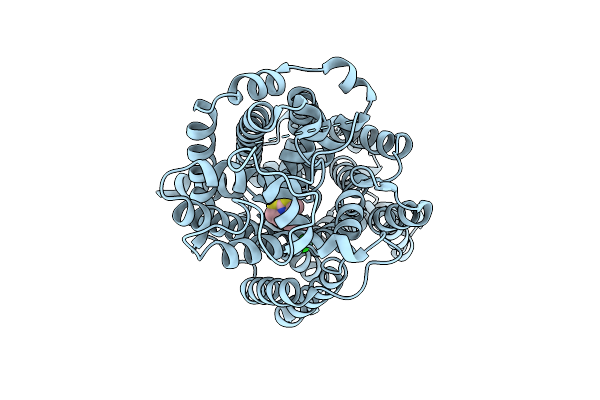 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: A1EBN, CL |
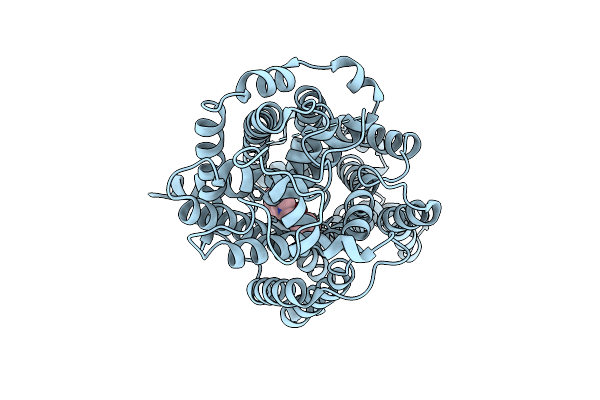 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: A1EBO, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-12-03 Classification: UNKNOWN FUNCTION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2025-12-03 Classification: UNKNOWN FUNCTION |
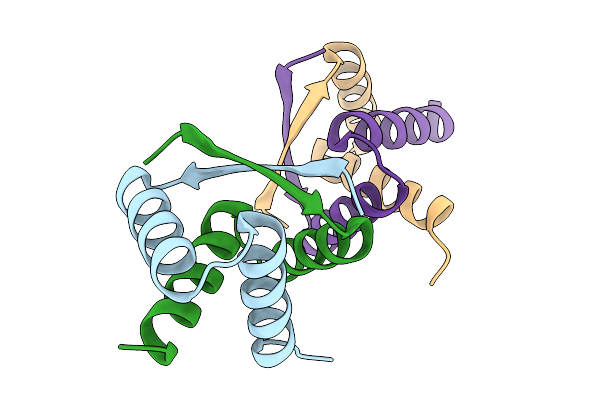 |
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-11-19 Classification: DNA BINDING PROTEIN |
 |
Organism: Escherichia phage
Method: ELECTRON MICROSCOPY Resolution:3.12 Å Release Date: 2025-11-12 Classification: VIRAL PROTEIN |
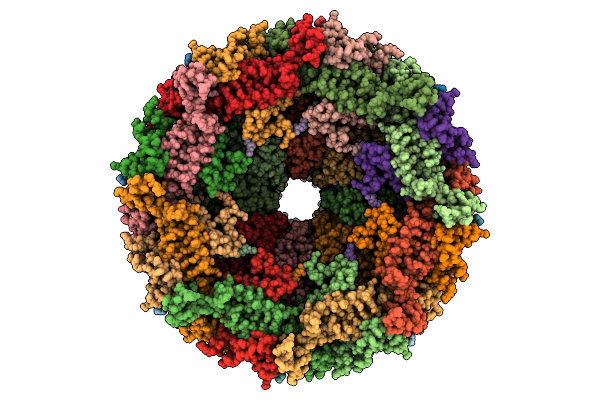 |
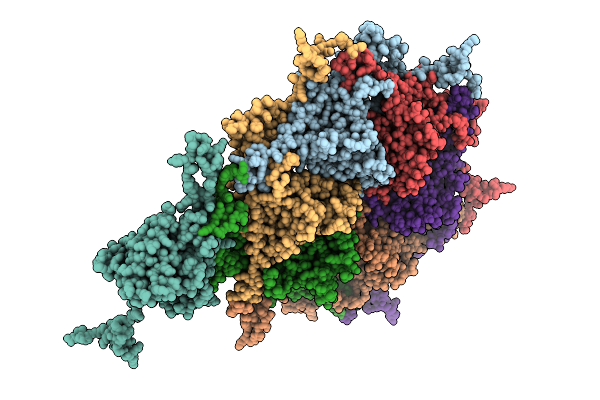
Organism: Escherichia phage
Method: ELECTRON MICROSCOPY Resolution:3.33 Å Release Date: 2025-11-12 Classification: VIRAL PROTEIN |
 |
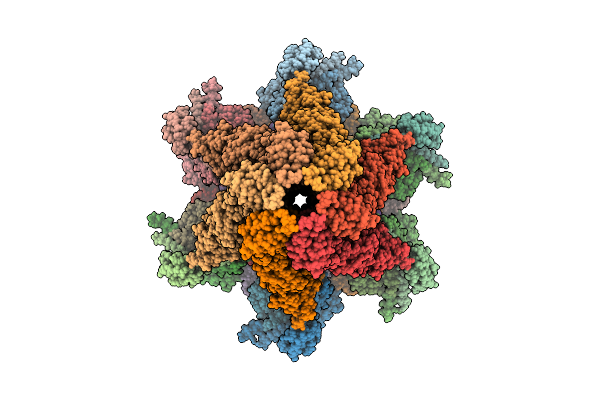
Organism: Escherichia phage
Method: ELECTRON MICROSCOPY Resolution:3.17 Å Release Date: 2025-11-12 Classification: VIRAL PROTEIN |
 |
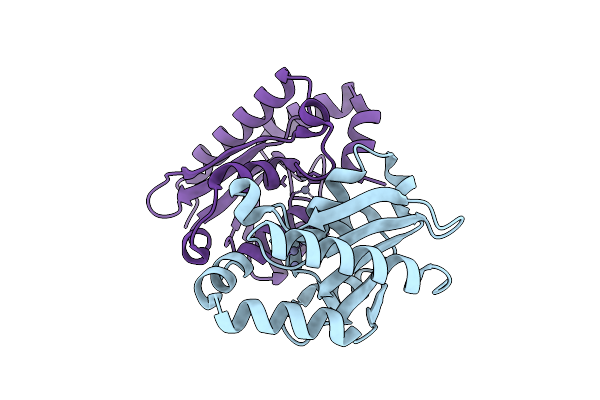
Organism: Escherichia phage
Method: ELECTRON MICROSCOPY Resolution:3.11 Å Release Date: 2025-11-12 Classification: VIRAL PROTEIN |
 |
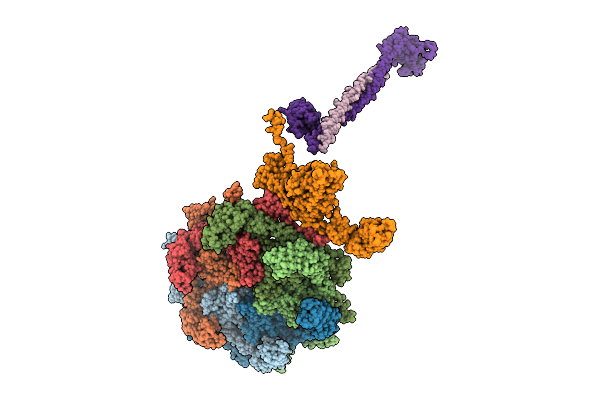
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2025-10-22 Classification: ANTITUMOR PROTEIN Ligands: ZN |
 |
Organism: Escherichia phage p1
Method: ELECTRON MICROSCOPY Resolution:3.60 Å Release Date: 2025-10-22 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia phage p1
Method: ELECTRON MICROSCOPY Resolution:3.50 Å Release Date: 2025-10-22 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia phage p1
Method: ELECTRON MICROSCOPY Resolution:3.60 Å Release Date: 2025-10-22 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia phage p1
Method: ELECTRON MICROSCOPY Resolution:3.70 Å Release Date: 2025-10-22 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia phage p1
Method: ELECTRON MICROSCOPY Resolution:4.00 Å Release Date: 2025-10-22 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia phage p1
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: VIRAL PROTEIN |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH4 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH5 |
 |
Organism: Human immunodeficiency virus 1
Method: X-RAY DIFFRACTION Resolution:3.36 Å Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: A1CH6 |

