Search Count: 28
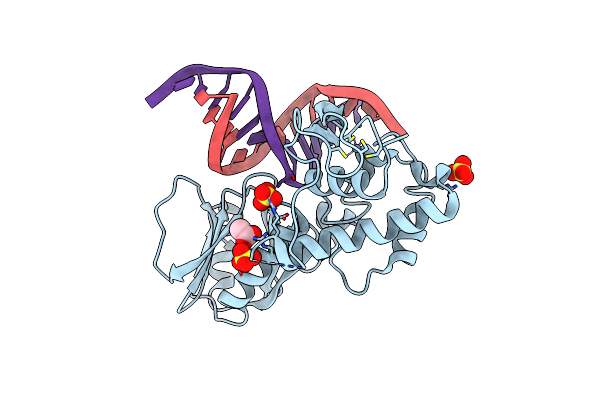 |
Crystal Structure Of The Dna Repair Enzyme Endonuclease-Viii (Nei) From E. Coli (E2Q) In Complex With Ap-Site Containing Dna Substrate
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-01-30 Classification: HYDROLASE Ligands: SO4, ZN, ACT |
 |
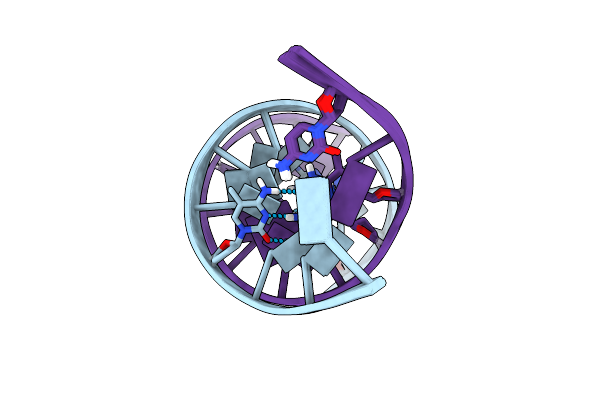
Solution Structure Of A Dna Dodecamer With 5-Methylcytosine At The 3Rd And 9Th Position
|
 |
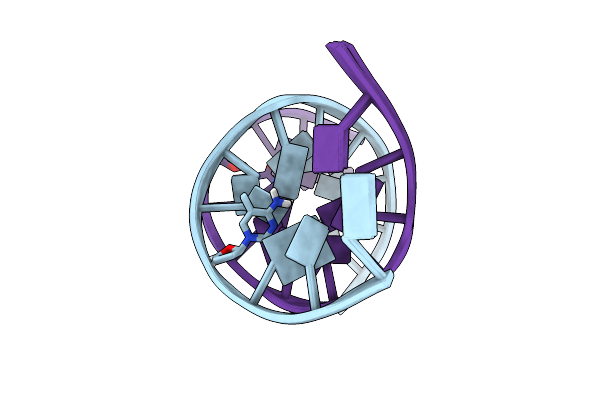
Solution Structure Of A Dna Dodecamer With 5-Methylcytosine At The 3Rd And 9Th Position And 8-Oxoguanine At The 4Th Position
|
 |
Solution Structure Of A Dna Dodecamer With 5-Methylcytosine At The 3Rd And 8-Oxoguanine At The 4Th Position
|
 |
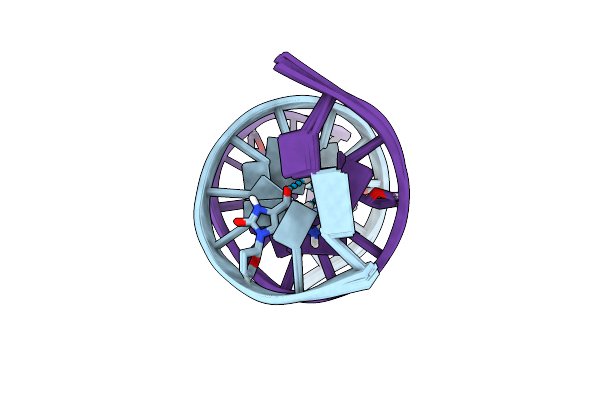
Solution Structure Of A Dna Dodecamer With 8-Oxoguanine At The 4Th Position And 5-Methylcytosine At The 9Th Position
|
 |
Solution Structure Of A Dna Dodecamer With 5-Methylcytosine At The 3Rd Position And 8-Oxoguanine At The 10Th Position
|
 |
Solution Structure Of A Dna Dodecamer With 5-Methylcytosine At The 9Th Position And 8-Oxoguanine At The 10Th Position
|
 |
Solution Structure Of A Dna Dodecamer With 5-Methylcytosine At The 3Rd And 9Th Position And 8-Oxoguanine At The 10Th Position
|
 |
Solution Structure Of A Dna Dodecamer With 5-Methylcytosine At The 3Rd Position
|
 |
Solution Structure Of A Dna Dodecamer With 5-Methylcytosine At The 9Th Position
|
 |
 |
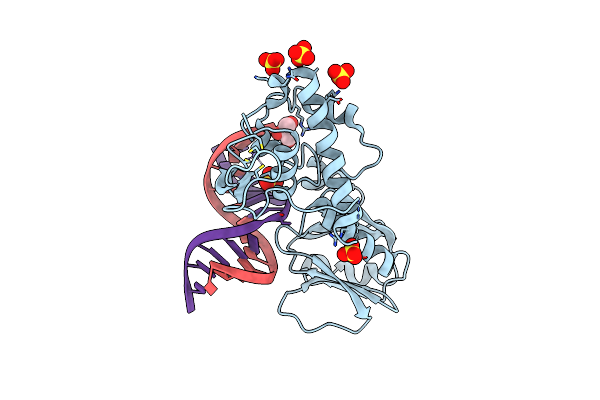 |
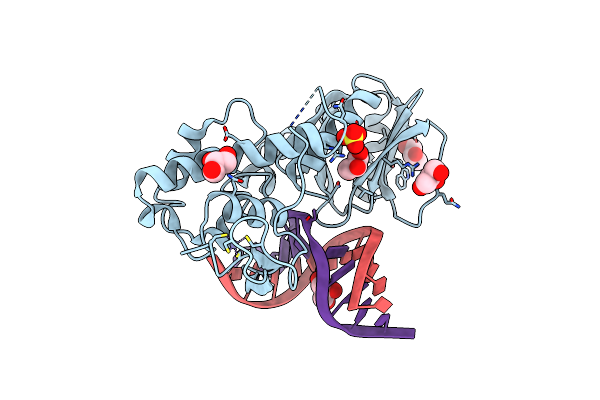
Crystal Structure Of The Dna Repair Enzyme Endonuclease-Viii (Nei) From E. Coli In Complex With Ap-Site Containing Dna Substrate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2008-02-26 Classification: HYDROLASE/DNA Ligands: SO4, ZN, GOL |
 |
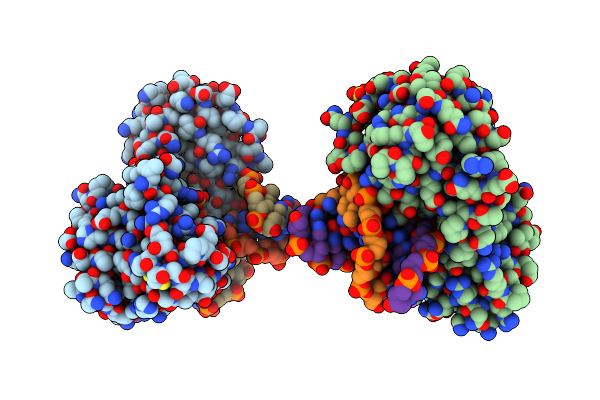
Crystal Structure Of The Dna Repair Enzyme Endonuclease-Viii (Nei) From E. Coli (R252A) In Complex With Ap-Site Containing Dna Substrate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2008-02-26 Classification: HYDROLASE/DNA Ligands: GOL, ZN, SO4 |
 |
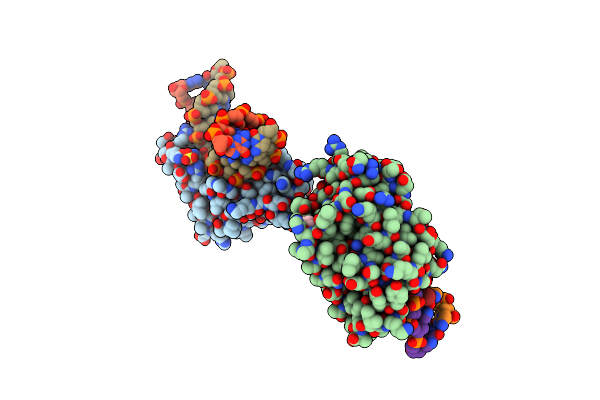
Crystal Structure Of The Dna Repair Enzyme Endonuclease-Viii (Nei) From E. Coli (E2Q) In Complex With Ap-Site Containing Dna Substrate
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-02-26 Classification: HYDROLASE/DNA |
 |
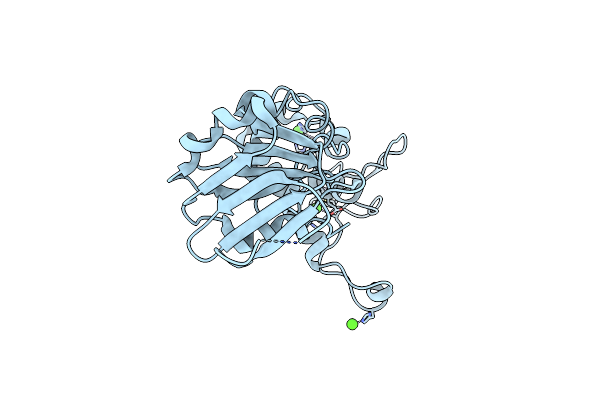
Crystal Structure Of T4 Pyrimidine Dimer Glycosylase (T4-Pdg) Covalently Complexed With A Dna Substrate Containing Abasic Site
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2006-10-03 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Crystal Structure Of The Dna Repair Enzyme Endonuclease-Viii (Nei) From E. Coli: The Wt Enzyme At 2.8 Resolution.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2004-08-03 Classification: HYDROLASE Ligands: ZN, CA |
 |
Crystal Structure Of The Dna Repair Enzyme Endonuclease-Viii (Nei) From E. Coli: The R252A Mutant At 2.05 Resolution.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2004-08-03 Classification: HYDROLASE Ligands: ZN, MG, GOL |
 |
Crystal Structure Of The Dna Repair Enzyme Endonuclease-Viii (Nei) From E. Coli: The E2A Mutant At 2.3 Resolution.
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2004-08-03 Classification: HYDROLASE Ligands: ZN, MG, GOL |
 |
Crystal Structure Of The Core Fragment Of Muty From E.Coli At 1.2A Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2002-11-26 Classification: HYDROLASE Ligands: SO4, SF4, GOL |

