Search Count: 318
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: LIGASE Ligands: ZN |
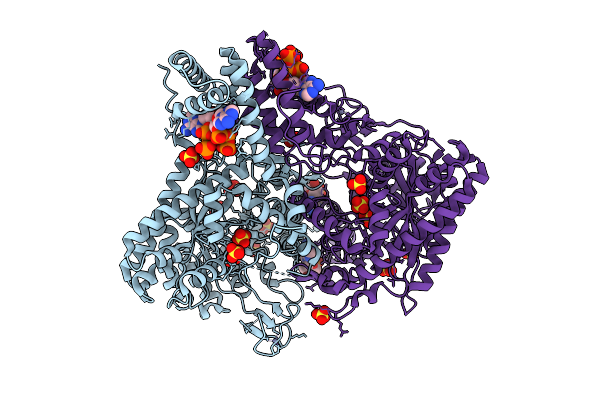 |
Crystal Structure Of S. Thermophilus Class Iii Ribonucleotide Reductase Bound To Datp
Organism: Streptococcus thermophilus
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: MG, DTP, ZN, SO4 |
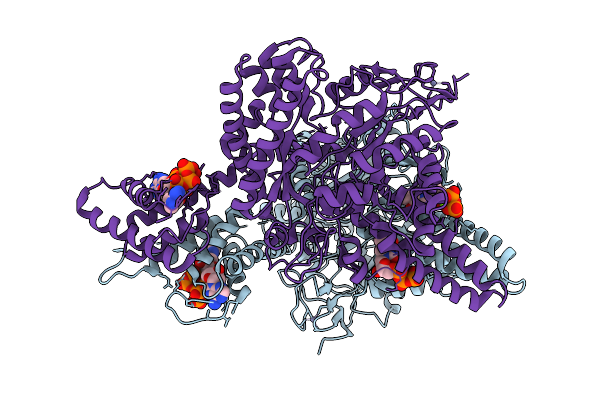 |
Organism: Streptococcus thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: TTP, DTP, MG, ZN |
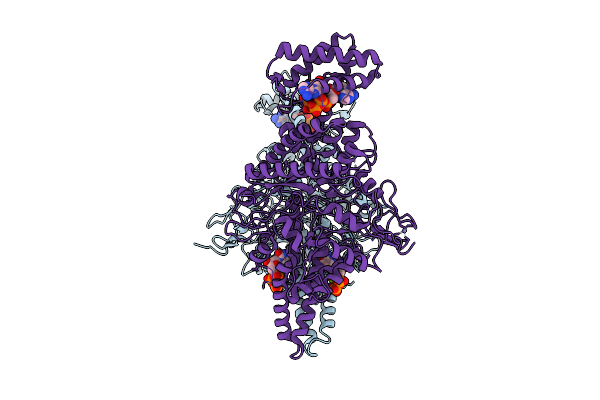 |
Organism: Streptococcus thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: ATP, MG, TTP, ZN |
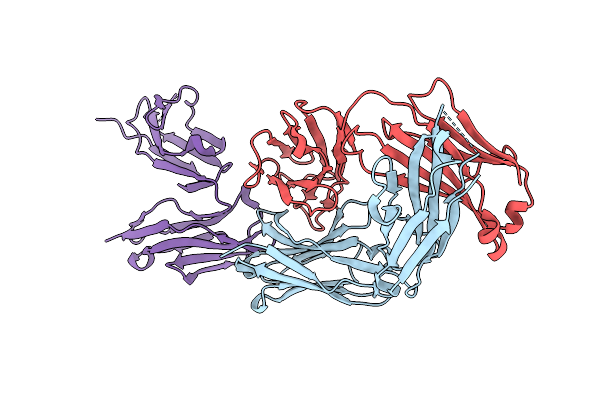 |
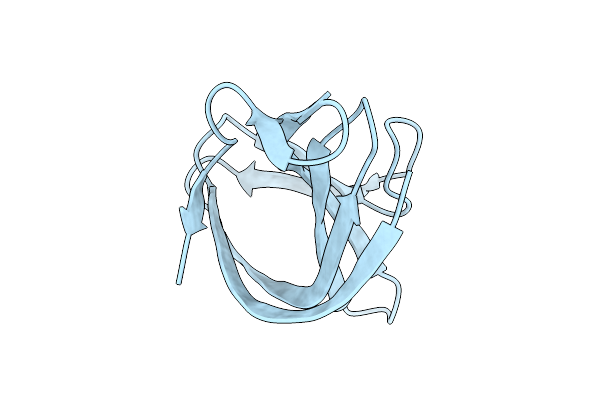
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
 |
Organism: Streptococcus phage philp081102
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: LIGASE |
 |
Crystal Structure Of The Polyester Hydrolase Leipzig 7 (Phl7) Mut3 Variant With Glycosylation By Expression In Pichia Pastoris (P_Phl7Mut3)
Organism: Compost metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of The Non-Glycosylated Polyester Hydrolase Leipzig 7 (Phl7) Mut3 Variant Expressed In Pichia Pastoris (P_Phl7Mut3_Ng)
Organism: Compost metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of The Polyester Hydrolase Leipzig 7 (Phl7) Mut3 Variant Expressed In E. Coli (E_Phl7Mut3)
Organism: Compost metagenome
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: HYDROLASE |
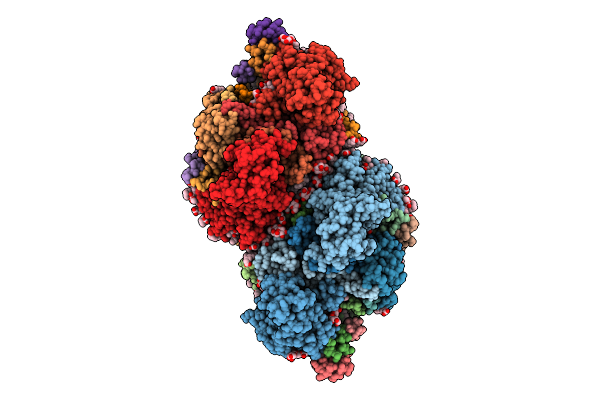 |
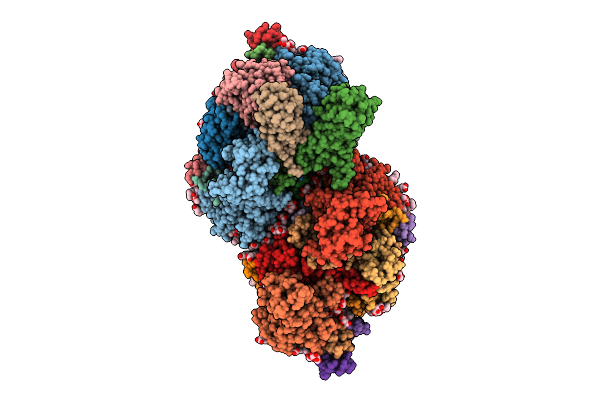
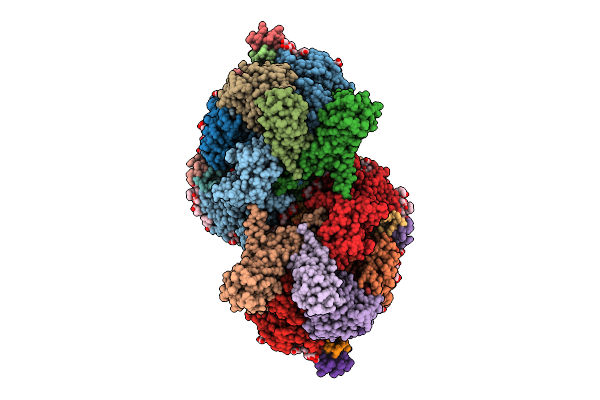
Organism: Thermosynechococcus vestitus bp-1
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: PHOTOSYNTHESIS Ligands: FE2, CL, CLA, PHO, BCR, SQD, PL9, LFA, PLM, BCT, LHG, LMG, LMT, DGD, HEM, RRX |
 |
 |
 |
Organism: Homo sapiens, Human coronavirus hku1 (isolate n1)
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: VIRAL PROTEIN Ligands: CA, NAG |
 |
Organism: Human coronavirus hku1 (isolate n1), Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ANTIVIRAL PROTEIN Ligands: NAG, GOL |
 |
Organism: Homo sapiens, Vicugna pacos
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ANTIVIRAL PROTEIN Ligands: CA, NAG, GOL, MLI |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ANTIVIRAL PROTEIN Ligands: MES, GOL, CA |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: ISOMERASE Ligands: MN, SO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: MEMBRANE PROTEIN Ligands: DXC |
 |
Structure Of Hku5 Spike C-Terminal Domain In Complex With Ace2 From Pipistrellus Abramus
Organism: Pipistrellus abramus, Pipistrellus bat coronavirus hku5
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: VIRAL PROTEIN Ligands: NAG |

