Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
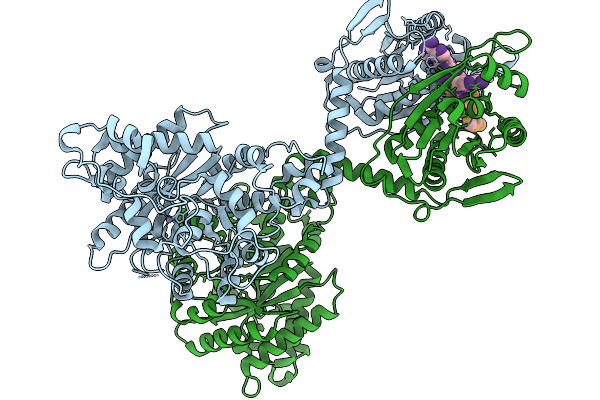 |
Crispr-Associated Deaminase Cad1 In Ca4 Bound Form, Symmetry Expanded Dimer, Refined Against A Composite Map
Organism: Thermoanaerobaculum aquaticum
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: SIGNALING PROTEIN Ligands: ZN |
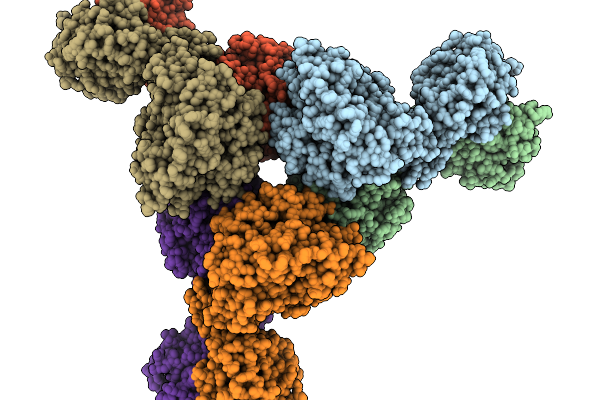 |
Organism: Thermoanaerobaculum aquaticum
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
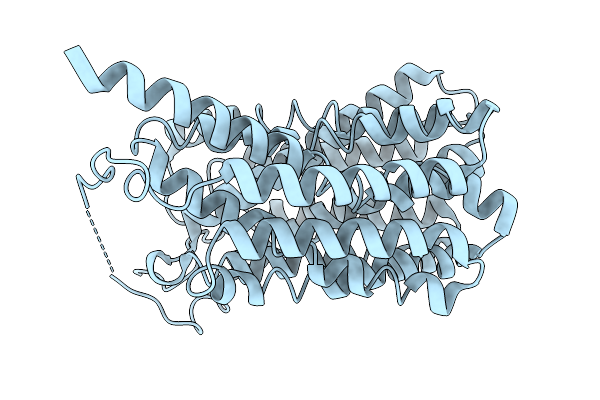 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN |
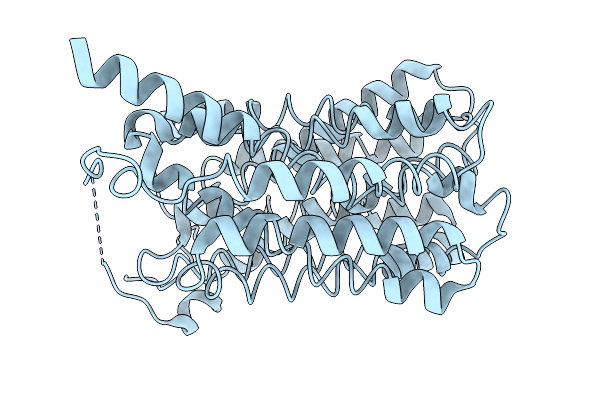 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN |
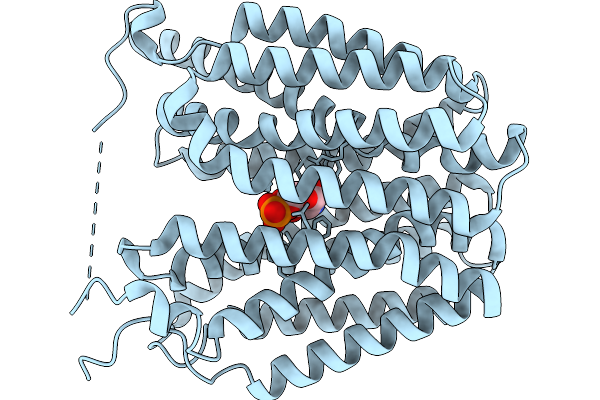 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: GLP |
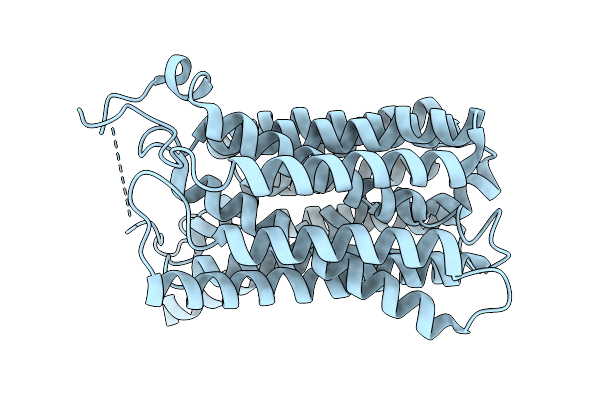 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN |
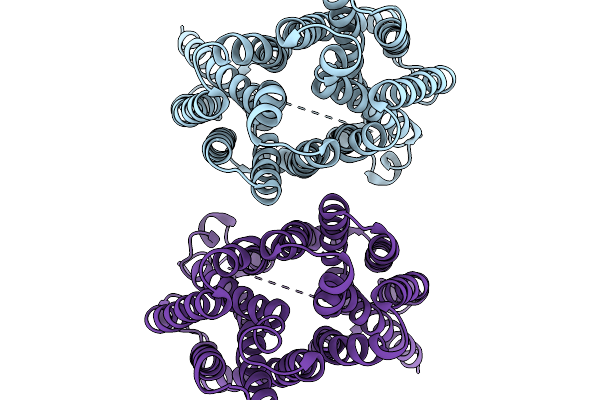 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN |
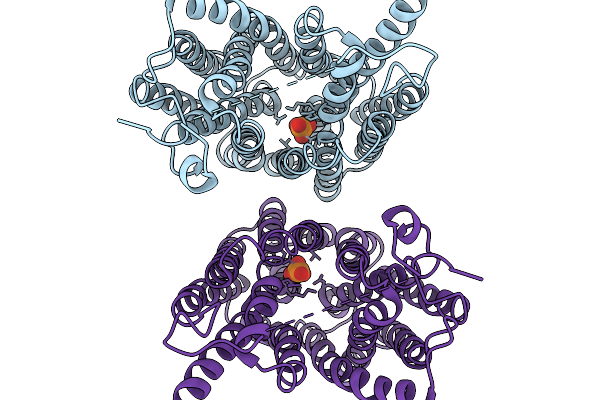 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: PO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: PO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: TRANSPORT PROTEIN Ligands: BG6 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: TRANSPORT PROTEIN Ligands: A1EG7 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: TRANSPORT PROTEIN Ligands: LMN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: TRANSPORT PROTEIN Ligands: LMN, PO4 |
 |
Organism: Vaccinia virus western reserve
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: GOL, CIT, A1I9D |
 |
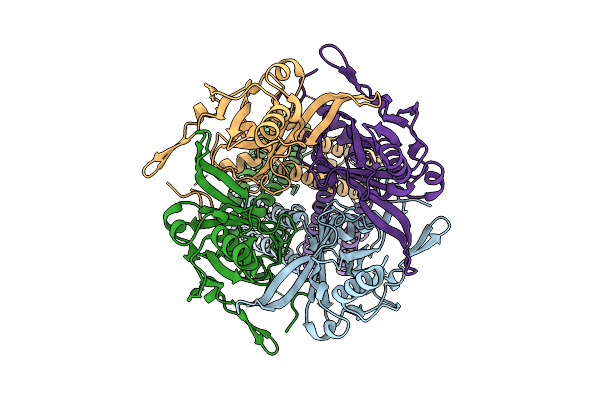
Cryoem Structure Of G Protein-Gated Inwardly Rectifying Potassium Channel Girk2 R92F (State 2)
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
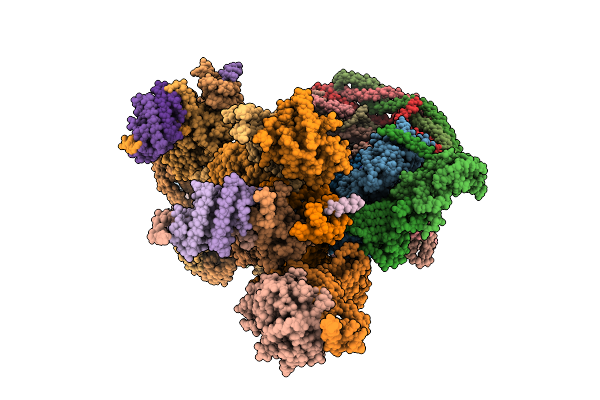
Cryoem Structure Of G Protein-Gated Inwardly Rectifying Potassium Channel 2 R92F (State 1)
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: MOTOR PROTEIN Ligands: ADP, ATP, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: UNKNOWN FUNCTION |