Search Count: 473
 |
Organism: Streptomyces klenkii
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2026-01-21 Classification: HYDROLASE |
 |
Structure Of Bfl1 In Complex With A Covalent Inhibitor, Alternative Series, Cmpd25
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2026-01-14 Classification: APOPTOSIS Ligands: A1JL2 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2026-01-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Severe fever with thrombocytopenia syndrome virus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2026-01-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Severe fever with thrombocytopenia syndrome virus
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2026-01-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
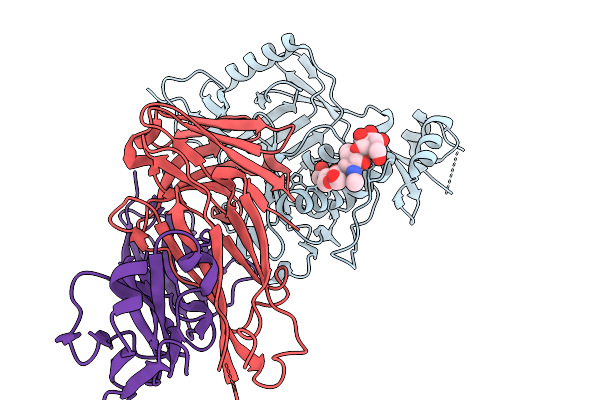 |
Organism: Homo sapiens, Severe fever with thrombocytopenia syndrome virus
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2026-01-07 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
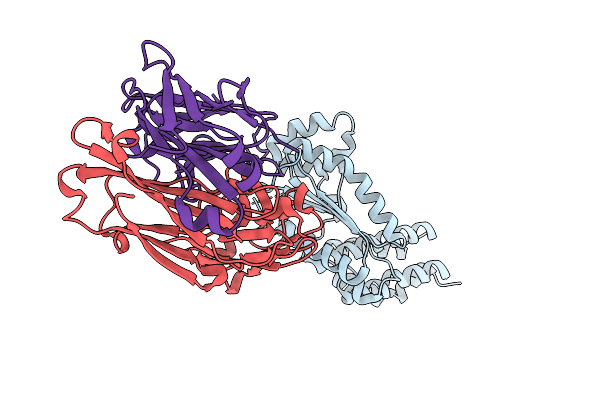
Crystal Structure Of Crimean-Congo Hemorrhagic Fever Virus Cap-Snatching Endonuclease
Organism: Orthonairovirus haemorrhagiae, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2025-12-31 Classification: HYDROLASE |
 |
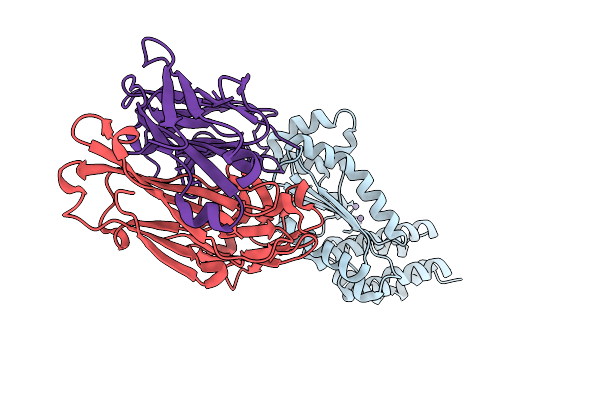
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-12-31 Classification: HYDROLASE |
 |
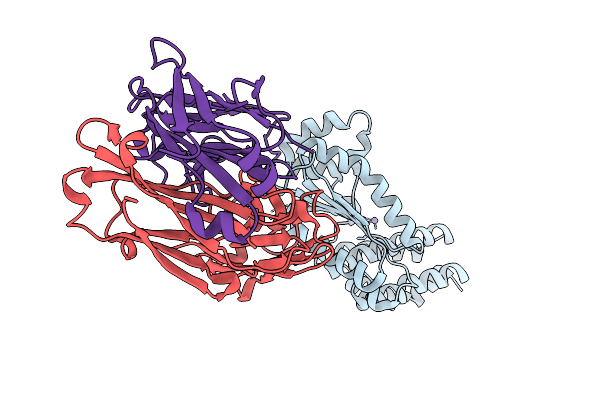
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With Manganese Ions
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease (E668A Mutant)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease (E682A Mutant)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease (D719A Mutant)
Organism: Mus musculus, Kasokero virus
Method: X-RAY DIFFRACTION Resolution:2.47 Å Release Date: 2025-12-31 Classification: HYDROLASE |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With 2,4-Dioxo-4-Phenylbutanoic Acid (Dpba)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN, XI7 |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With L-742,001
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN, 0N8 |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With Baloxavir Acid (Bxa)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN, E4Z |
 |
Organism: Henipavirus nipahense, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-11-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Henipavirus nipahense, Mesocricetus auratus
Method: ELECTRON MICROSCOPY Resolution:3.03 Å Release Date: 2025-11-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-10-15 Classification: TRANSCRIPTION Ligands: A1EKB |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2025-10-15 Classification: TRANSCRIPTION Ligands: A1EJ9 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2025-10-15 Classification: TRANSCRIPTION Ligands: A1EJ8 |

