Search Count: 172
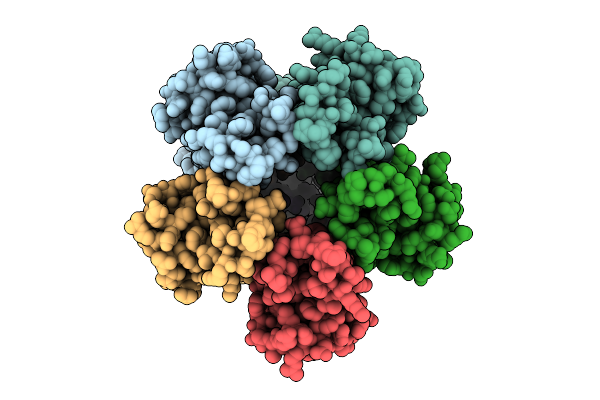 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
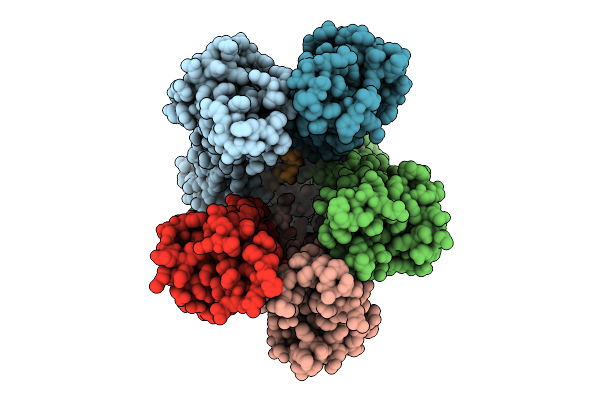 |
Organism: Escherichia coli str. k-12 substr. mg1655
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
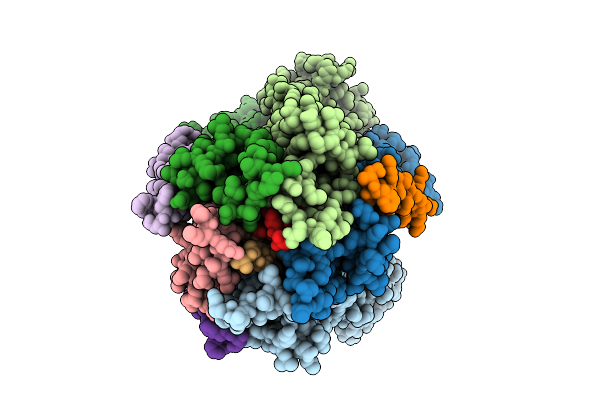 |
Organism: Escherichia coli str. k-12 substr. mg1655
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
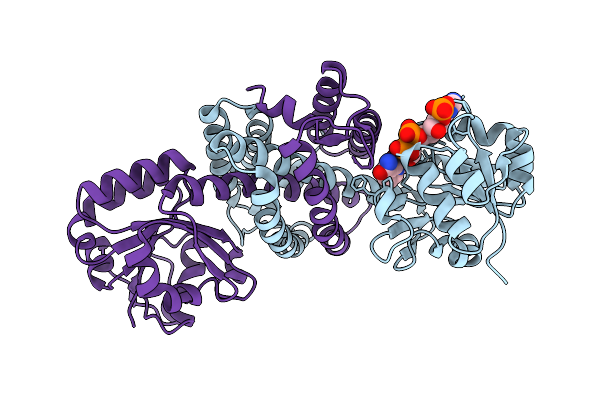 |
Structure Of Imine Reductase 361 From Micromonospora Sp. Mutant M125W/I127F/L179V/H250L
Organism: Micromonospora
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
Organism: Homo sapiens, Streptococcus pneumoniae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: SIGNALING PROTEIN/IMMUNE SYSTEM Ligands: SA0 |
 |
Organism: Homo sapiens, Streptococcus pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: SIGNALING PROTEIN Ligands: SA0 |
 |
Organism: Homo sapiens, Streptococcus pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: SIGNALING PROTEIN Ligands: SA0 |
 |
Organism: Homo sapiens, Streptococcus pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2025-04-02 Classification: SIGNALING PROTEIN Ligands: SA0 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN Ligands: PEG, A1IGD, CL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN Ligands: IOD, PG4 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2025-01-29 Classification: BIOSYNTHETIC PROTEIN Ligands: PG4, PEG, CL, PGE |
 |
Organism: Isochrysis galbana
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: PHOTOSYNTHESIS Ligands: CLA, KC2, DD6, LHG, LMG, A86, LMU, SQD, DGD, BCR, SF4, PQN |
 |
Organism: Alkalimonas delamerensis
Method: X-RAY DIFFRACTION Resolution:1.26 Å Release Date: 2024-12-18 Classification: HYDROLASE Ligands: B3P, TRS |
 |
The Substrate Binding Protein Of An Abc Transporter In Complex With Beta-1,4-Xylobiose
Organism: Vibrio sp. ea2
Method: X-RAY DIFFRACTION Release Date: 2024-12-11 Classification: TRANSPORT PROTEIN Ligands: GOL |
 |
The Substrate Binding Protein Of An Abc Transporter In Complex With Beta-1,3-Xylobiose
Organism: Vibrio sp. ea2
Method: X-RAY DIFFRACTION Release Date: 2024-12-11 Classification: TRANSPORT PROTEIN |
 |
The Substrate Binding Protein Of An Abc Transporter In Complex With Beta-1,3-Xylotriose
Organism: Vibrio sp. ea2
Method: X-RAY DIFFRACTION Release Date: 2024-12-11 Classification: TRANSPORT PROTEIN |
 |
Organism: Alkalimonas delamerensis
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-12-11 Classification: HYDROLASE |
 |
Organism: Synthetic construct, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-09-18 Classification: STRUCTURAL PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-08-07 Classification: STRUCTURAL PROTEIN |

