Search Count: 20
 |
Organism: Acinetobacter bereziniae niph 3
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2023-10-04 Classification: LYASE Ligands: ZN, LNI |
 |
Organism: Candidatus pelagibacter ubique htcc1062
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2023-10-04 Classification: LYASE Ligands: MN, LNI |
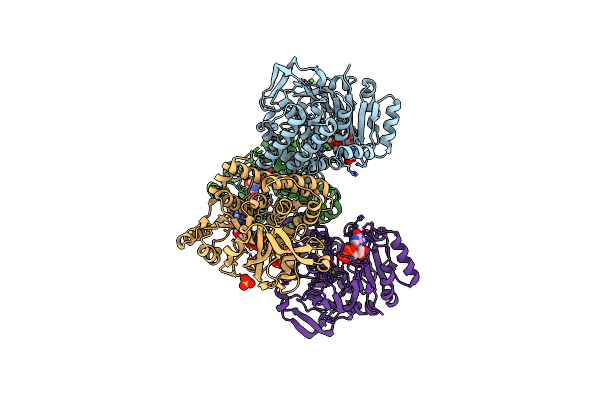 |
Structural Insight Into A Metal-Dependent Mutase Mtdl Revealing An Arginine Residue Covalently Mediated Interconversion Between Nucleotide-Based Furanose And Pyranose
Organism: Marinactinospora thermotolerans
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-07-26 Classification: BIOSYNTHETIC PROTEIN Ligands: GDP, MG, SO4 |
 |
Structural Insight Into A Metal-Dependent Mutase Mtdl Revealing An Arginine Residue Covalently Mediated Interconversion Between Nucleotide-Based Furanose And Pyranose
Organism: Marinactinospora thermotolerans
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2023-07-26 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4 |
 |
Organism: Vibrio splendidus
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-11-16 Classification: LYASE Ligands: SO4 |
 |
Organism: Neopyropia yezoensis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-08-17 Classification: LYASE |
 |
Organism: Vibrio pelagius
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-08-17 Classification: LYASE Ligands: GOL |
 |
Organism: Neopyropia yezoensis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-08-17 Classification: LYASE |
 |
Organism: Nisaea denitrificans dsm 18348
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-06-01 Classification: TRANSFERASE Ligands: SAM |
 |
Organism: Nisaea denitrificans dsm 18348
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2022-06-01 Classification: TRANSFERASE Ligands: SAH, H9L, TRS |
 |
Organism: Glycine max
Method: ELECTRON MICROSCOPY Release Date: 2022-03-16 Classification: MEMBRANE PROTEIN |
 |
Organism: Psychrobacter sp.
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-05-19 Classification: LYASE Ligands: ATP, SO4 |
 |
Organism: Myroides profundi
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2020-07-08 Classification: FLAVOPROTEIN Ligands: NAP, FAD |
 |
Organism: Ruegeria lacuscaerulensis (strain dsm 11314 / kctc 2953 / iti-1157)
Method: X-RAY DIFFRACTION Resolution:2.23 Å Release Date: 2019-07-03 Classification: LIGASE Ligands: ADP |
 |
Structure Of 3-Methylmercaptopropionate Coa Ligase Mutant K523A In Complex With Amp And Mmpa
Organism: Ruegeria lacuscaerulensis (strain dsm 11314 / kctc 2953 / iti-1157)
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2019-07-03 Classification: LIGASE Ligands: AMP, B3P, A8C |
 |
Organism: Roseovarius nubinhibens ism
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-07-03 Classification: OXIDOREDUCTASE Ligands: B3P |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2012-01-18 Classification: TRANSCRIPTION Ligands: ZN, CA |
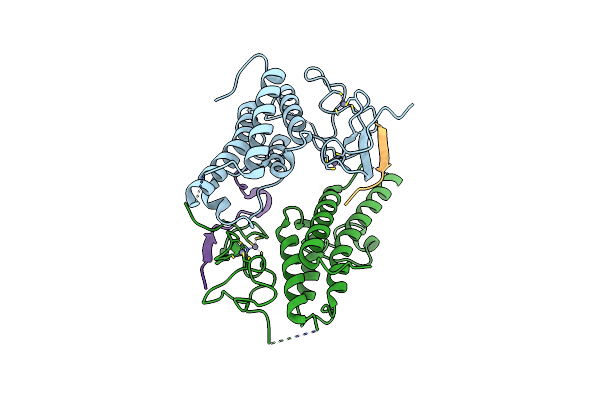 |
Crystal Structure Of The Complex Of Trim33 Phd-Bromo And H3(1-20)K9Me3K14Ac Histone Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2012-01-18 Classification: TRANSCRIPTION Ligands: ZN |
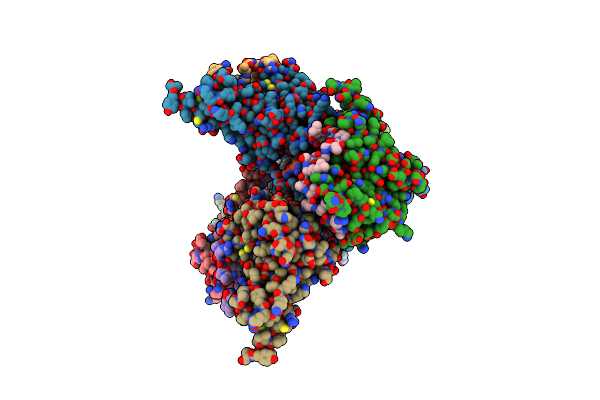 |
Crystal Structure Of The Complex Of Trim33 Phd-Bromo And H3(1-22)K9Me3K14Ack18Ac Histone Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-01-18 Classification: TRANSCRIPTION Ligands: ZN |
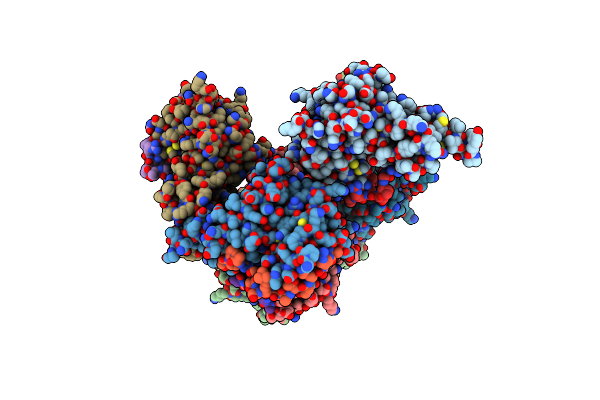 |
Crystal Structure Of The Complex Of Trim33 Phd-Bromo And H3(1-28)K9Me3K14Ack18Ack23Ac Histone Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2012-01-18 Classification: TRANSCRIPTION Ligands: ZN |

