Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Pectobacterium atrosepticum scri1043
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Pectobacterium atrosepticum scri1043, Thiocystis violascens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Pectobacterium atrosepticum scri1043, Thiocystis violascens dsm 198
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RNA BINDING PROTEIN/RNA |
 |
Cyro-Em Structure Of Prefusion Rsv Fusion Glycoprotein In Complex With Ziresovir And Motavizumab Fab
Organism: Human respiratory syncytial virus a2, Tequatrovirus t4, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: A1EV1 |
 |
Organism: Pectobacterium atrosepticum scri1043, Thiocystis violascens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: IMMUNE SYSTEM/RNA |
 |
Organism: Pectobacterium atrosepticum scri1043, Thiocystis violascens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: IMMUNE SYSTEM/RNA |
 |
Comparative Analysis Of Functions And Catalytic Mechanisms Of Methyltransferases Involved In Anthracycline Biosynthesis
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: TRANSFERASE Ligands: SAH, A1L3V |
 |
Comparative Analysis Of Functions And Catalytic Mechanisms Of Methyltransferases Involved In Anthracycline Biosynthesis
Organism: Streptomyces coeruleorubidus
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: TRANSFERASE Ligands: A1EI6, SAH |
 |
Crystal Structure Of A Transaminase Pata From Pseudonocardia Ammonioxydans In Complex With Plp And Llp
Organism: Pseudonocardia ammonioxydans
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: PLP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: A1EBN, CL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: A1EBO, CL |
 |
Organism: Human t-cell leukemia virus type i
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN |
 |
High-Resolution Analysis Of The Human T-Cell Leukemia Virus Capsid Protein Reveals Insights Into Immature Particle Morphology
Organism: Human t-cell leukemia virus type i
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRUS LIKE PARTICLE Ligands: IHP |
 |
Crystal Structure Of Crebbp Histone Acetyltransferase Domain In Complex With Acetyl-Coenzyme A
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: ZN, ACO |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
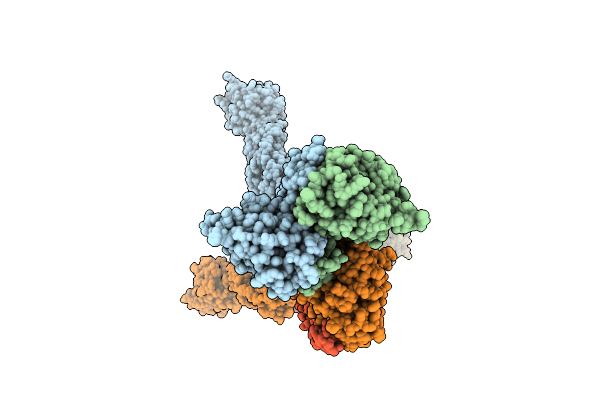
Cryo-Em Structure Of The Full-Length Pag-Bound Btn2A1-Btn3A1-Btn3A2 Complex
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: H6P |
 |
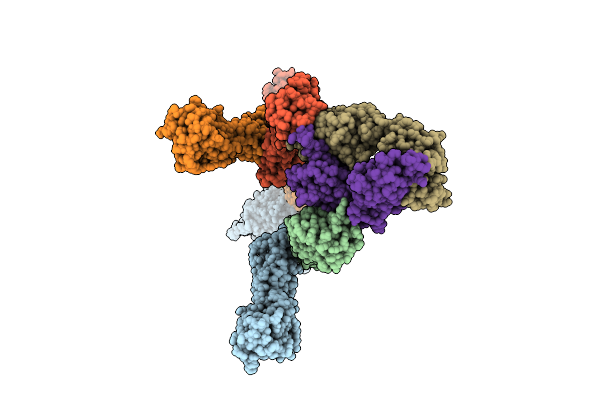
Cryo-Em Structure Of The Full Length Pag-Bound Btn2A1-Btn3A1-Btn3A2 In Complex With Vgamma9-Vdelta2 Tcr (Mop Genotype)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: H6P |
 |
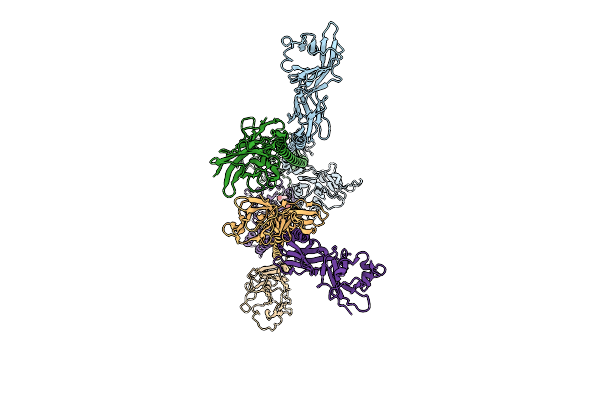
Cryo-Em Structure Of Pag-Bound Btn2A1-Btn3A1-Btn3A3 In Complex With Vgamma9-Vdelta2 Tcr (G115 Genotype)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: H6P |
 |
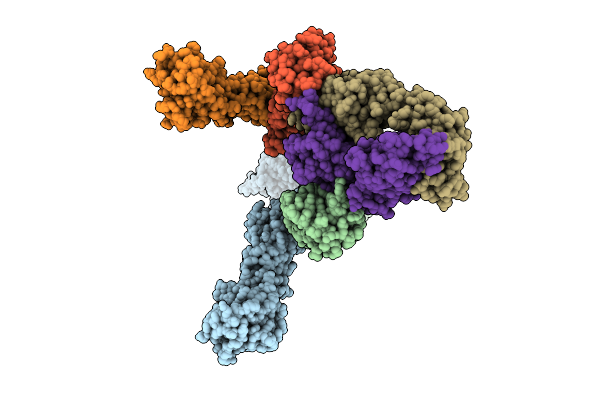
Cryo-Em Structure Of The Full-Length Pag-Bound Btn2A1-Btn3A1-Btn3A3 Complex
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: H6P |
 |
Cryo-Em Structure Of The Full Length Pag-Bound Btn2A1-Btn3A1-Btn3A2 In Complex With Vgamma9-Vdelta2 Tcr (G115 Genotype)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM Ligands: H6P |