Search Count: 522
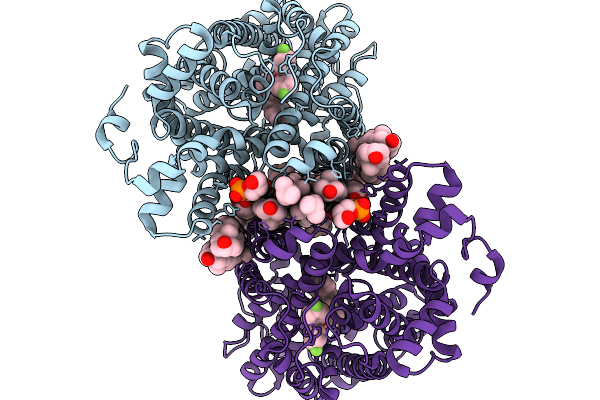 |
Cryo-Em Structure Of Lipid-Mediated Dimer Of Human Norepinephrine Transporter Net In The Presence Of Vanoxerine In An Inward-Open State At Resolution Of 2.52 Angstrom
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSPORT PROTEIN Ligands: CLR, A1D5S, A1EFM |
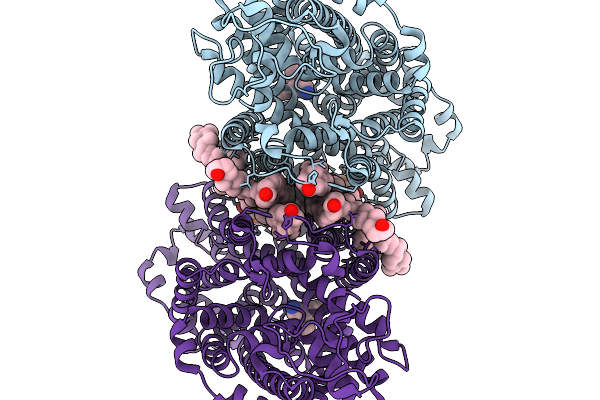 |
Cryo-Em Structure Of Lipid-Mediated Human Norepinephrine Transporter Net In The Presence Of Levomilnacipran In An Inward-Open State At Resolution Of 2.46 Angstrom.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSPORT PROTEIN Ligands: F0F, CLR, A1EFM |
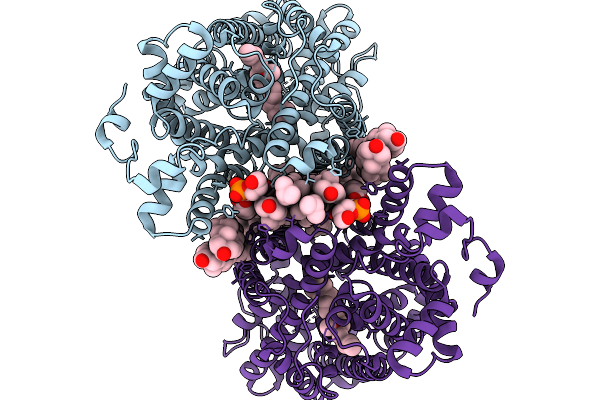 |
Cryo-Em Structure Of Lipid-Mediated Dimer Of Human Norepinephrine Transporter Net In The Presence Of The F3288-0031 In An Inward-Open State At Resolution Of 3.1 Angstrom
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: TRANSPORT PROTEIN Ligands: A1EFR, CLR, A1EFM |
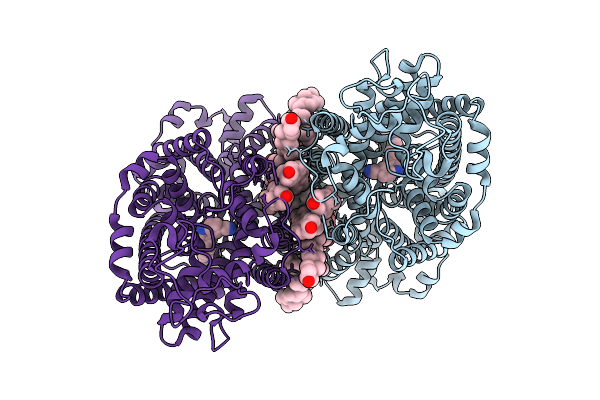 |
Cryo-Em Structure Of Lipid-Mediated Dimer Of Human Norepinephrine Transporter Net In The Presence Of The Antidepressant Vilazodone In An Inward-Open State At Resolution Of 2.44 Angstrom.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: PROTON TRANSPORT Ligands: YG7, CLR, A1EFM |
 |
Organism: Burkholderia cenocepacia h111
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: METAL BINDING PROTEIN Ligands: FE, ACY, FMT, NA |
 |
Structure Of The Recombinant Structure Of The Subunit Of Allophycocyanin (Apc) And The Formate Dehydrogenase (Fdh)
Organism: Candida boidinii, Picosynechococcus sp. pcc 7003
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: PHOTOSYNTHESIS Ligands: CYC |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSLOCASE Ligands: ZN, AD9 |
 |
Organism: Pseudomonas aeruginosa
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: TRANSLOCASE Ligands: ZN, AD9, LMT |
 |
Organism: Azotobacter vinelandii
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: LIGASE |
 |
Organism: Glycine max
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: PLANT PROTEIN |
 |
Organism: Glycine max
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: PLANT PROTEIN |
 |
Organism: Glycine max
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: PLANT PROTEIN |
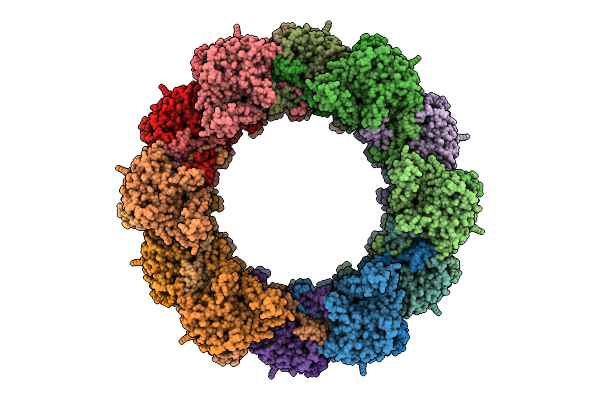 |
Azotobacter Vinelandii Extended Type Vi Secretion System Sheath Tube Complex
Organism: Azotobacter vinelandii
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: TRANSPORT PROTEIN |
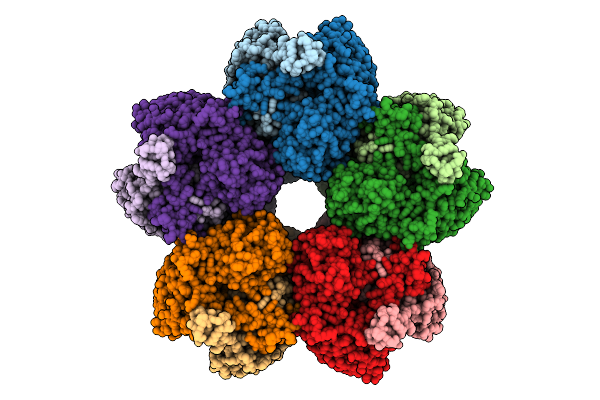 |
Organism: Azotobacter vinelandii
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: ISOMERASE |
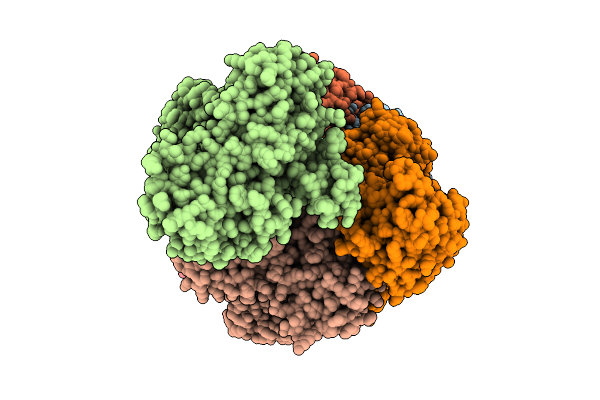 |
Cryoem Structure Of A Filamentous Soluble Pyridine Nucleotide Transhydrogenase
Organism: Azotobacter vinelandii
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: FLAVOPROTEIN Ligands: FAD |
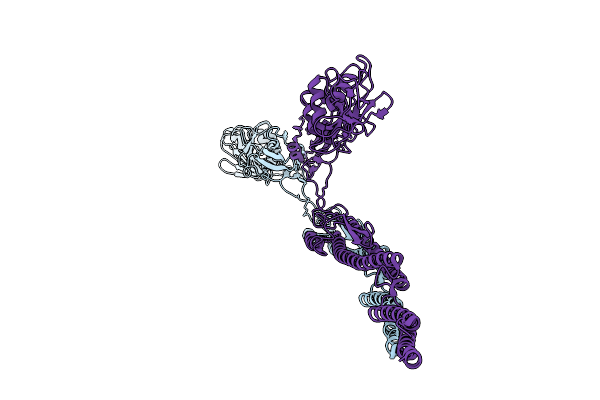 |
Organism: Azotobacter vinelandii
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: STRUCTURAL PROTEIN |
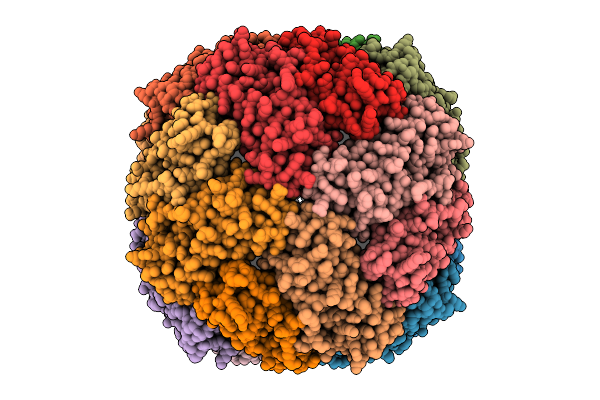 |
Organism: Azotobacter vinelandii
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: METAL BINDING PROTEIN Ligands: HEM |
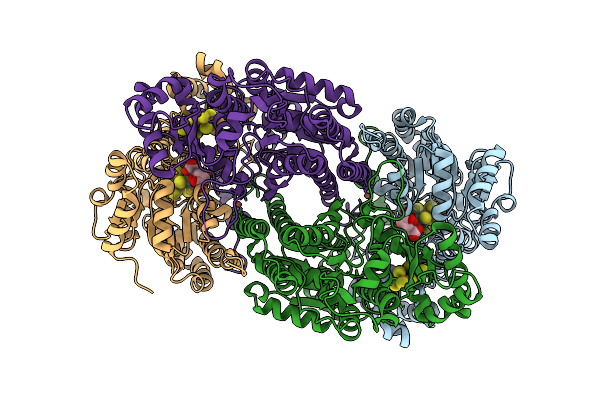 |
Organism: Azotobacter vinelandii
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: OXIDOREDUCTASE Ligands: ICS, HCA, CLF, FE |
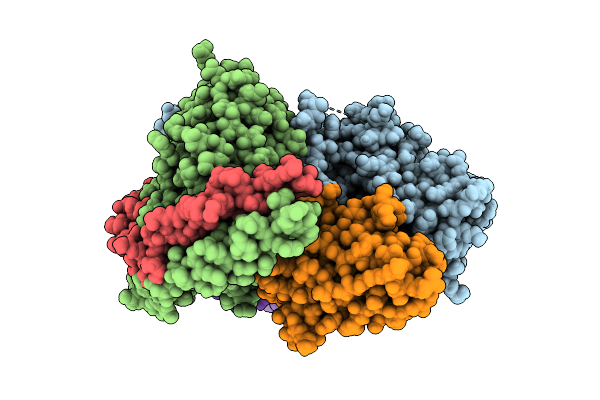 |
Cryo-Em Structure Of The Proton-Sensing Gpcr (Gpr4)-Gs Protein Complex At Ph 6.5
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: SIGNALING PROTEIN Ligands: CLR, A1L35 |
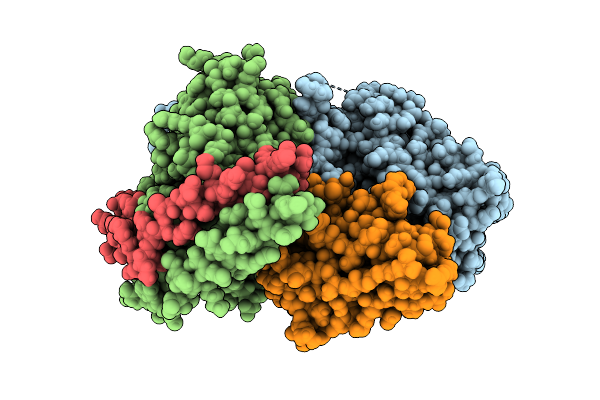 |
Cryo-Em Structure Of The Proton-Sensing Gpcr (Gpr4)-Gq Protein Complex At Ph 7.4
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: SIGNALING PROTEIN Ligands: CLR, A1L35 |

