Search Count: 25
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: PROTEIN FIBRIL Ligands: A1EFK |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: PROTEIN FIBRIL Ligands: A1EFL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: PROTEIN FIBRIL Ligands: A1EFL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: PROTEIN FIBRIL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: DE NOVO PROTEIN Ligands: ZN, ACT, FMT, GOL |
 |
Organism: Frankliniella occidentalis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-12-04 Classification: LIPID BINDING PROTEIN |
 |
Crystal Structure Of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed With Nadp+ And Fad In Space Group Of P21221
Organism: Thermocrispum municipale
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2022-07-06 Classification: FLAVOPROTEIN Ligands: FAD, NAP, PEG, EDO, NA |
 |
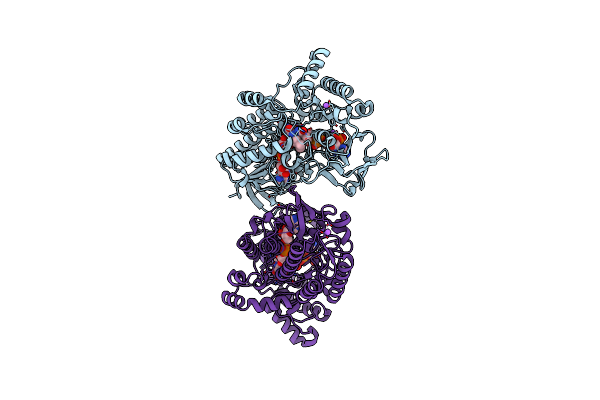
Crystal Structure Of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed With Nadp+ And Fad In Space Group Of C2221
Organism: Thermocrispum municipale
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2022-07-06 Classification: FLAVOPROTEIN Ligands: FAD, NAP, EDO, GOL, NA |
 |
Crystal Structure Of Cyclohexanone Monooxygenase From T. Municipale Mutant L437T Complexed With Nadp+ And Fad In Space Group Of P1211
Organism: Thermocrispum municipale
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2022-07-06 Classification: FLAVOPROTEIN Ligands: FAD, NAP, NA |
 |
Organism: Synthetic
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2019-03-13 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-02-20 Classification: DE NOVO PROTEIN Ligands: MPD, PO4, CO3 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.55 Å Release Date: 2018-08-15 Classification: DE NOVO PROTEIN Ligands: MPD, ZN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-08-15 Classification: DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2018-08-15 Classification: DE NOVO PROTEIN Ligands: HOH |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-08-15 Classification: DE NOVO PROTEIN Ligands: FMT, MPD |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-08-15 Classification: DE NOVO PROTEIN Ligands: PO4, MPD |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-08-15 Classification: DE NOVO PROTEIN Ligands: GOL |
 |
De Novo Design Of Polynuclear Transition Metal Clusters In Helix Bundles-4Eh1
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-01-03 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
De Novo Design Of Polynuclear Transition Metal Clusters In Helix Bundles-4Eh2
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-01-03 Classification: DE NOVO PROTEIN Ligands: ZN |
 |
De Novo Design Of Polynuclear Transition Metal Clusters In Helix Bundles-4Dh1
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-01-03 Classification: DE NOVO PROTEIN Ligands: ZN, GOL |

