Search Count: 972
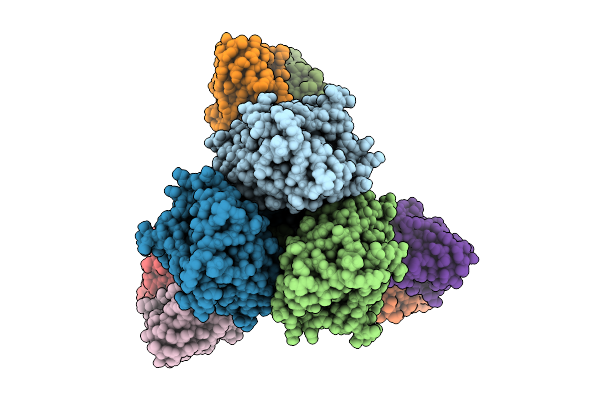 |
Organism: Alphainfluenzavirus influenzae, Macaca mulatta
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: VIRAL PROTEIN |
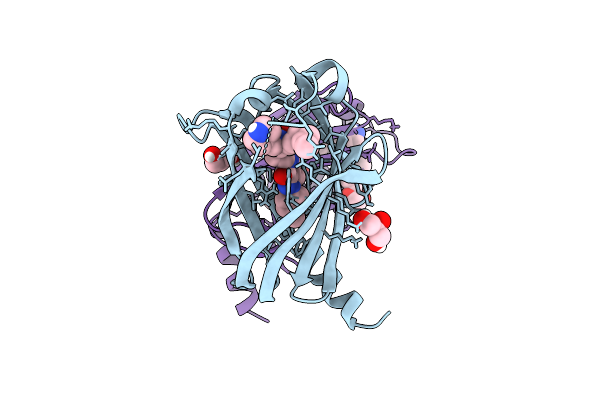 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2025-12-24 Classification: LIPID BINDING PROTEIN Ligands: A1IWC, GOL, PEG |
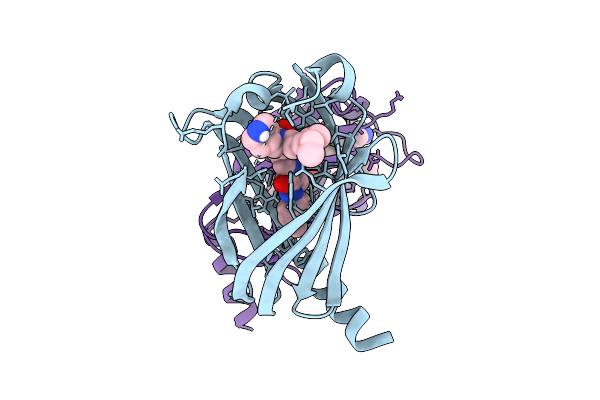 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-12-24 Classification: LIPID BINDING PROTEIN Ligands: A1IWD |
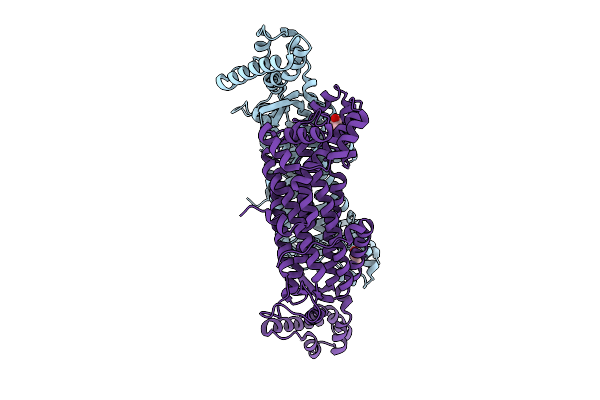 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-12-24 Classification: LYASE Ligands: GOL |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2025-12-24 Classification: LYASE Ligands: GOL |
 |
Organism: Vaccinia virus western reserve
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: GOL, CIT, A1I9D |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.17 Å Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.29 Å Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
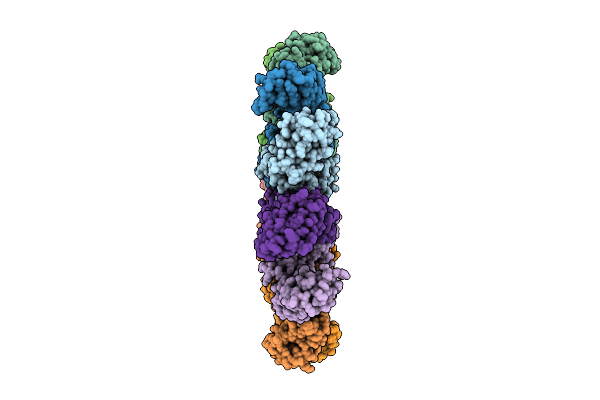 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.75 Å Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
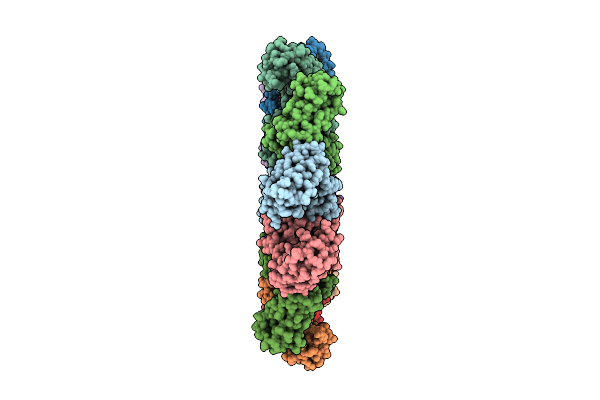 |
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Resolution:3.29 Å Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
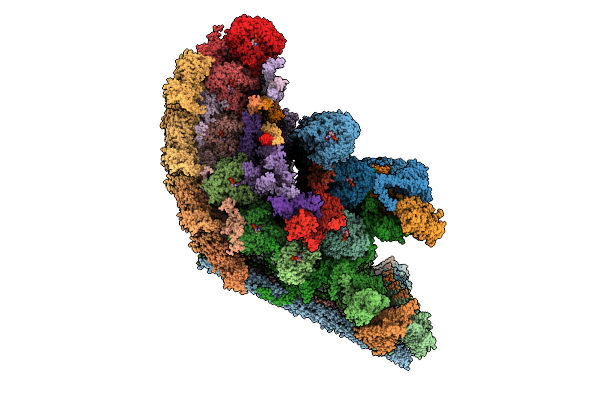 |
Organism: Toxoplasma gondii
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: STRUCTURAL PROTEIN Ligands: GTP, MG, GDP |
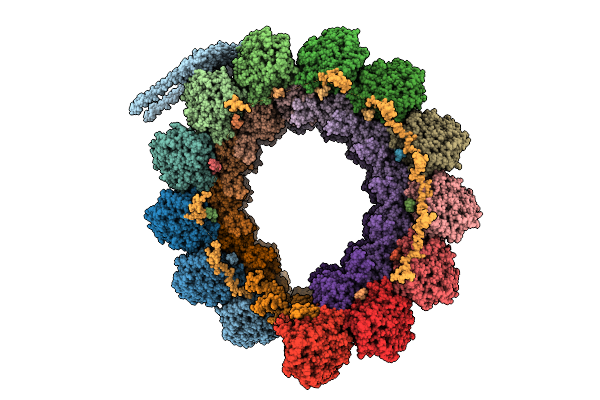 |
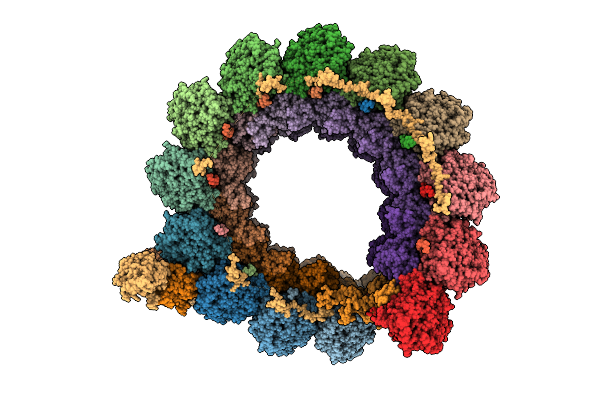
Cryo-Em Structure Of Intraconoidal Microtubule 2 (Icmt2) From Toxoplasma Gondii (8-Nm Repeat)
Organism: Toxoplasma gondii
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: STRUCTURAL PROTEIN Ligands: GTP, MG, GDP |
 |
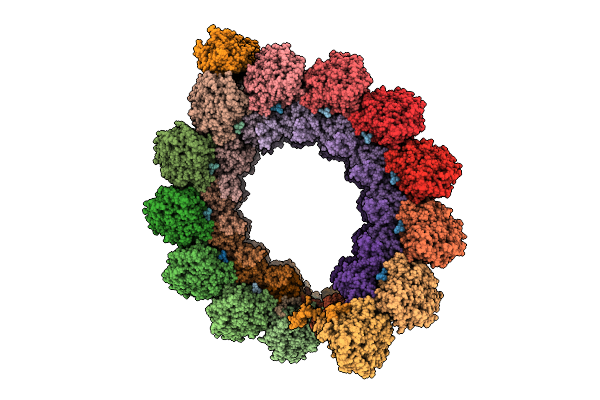
Cryo-Em Structure Of Intraconoidal Microtubule 1 (Icmt1) From Toxoplasma Gondii (8-Nm Repeat)
Organism: Toxoplasma gondii
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: STRUCTURAL PROTEIN Ligands: GTP, MG, GDP |
 |
Cryo-Em Structure Of The Apical Region Of Subpellicular Microtubule (Spmt) From Toxoplasma Gondii (8-Nm Repeat)
Organism: Toxoplasma gondii
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: STRUCTURAL PROTEIN Ligands: GDP, GTP, MG |
 |
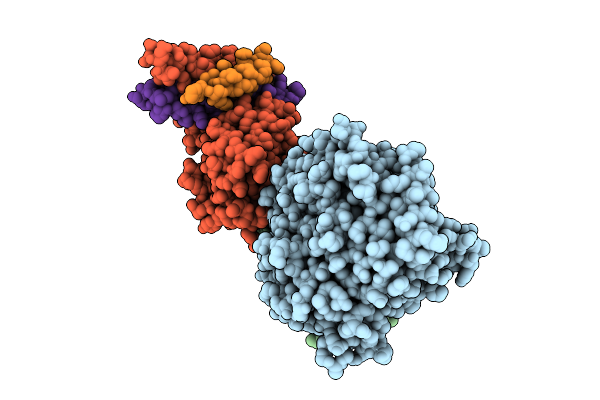
Organism: Toxoplasma gondii
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: STRUCTURAL PROTEIN |
 |
Symmetry-Expanded Reconstruction Of Augmin T-Ii Bonsai On The Gtpgammas Microtubule
Organism: Xenopus laevis, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: CELL CYCLE Ligands: GTP, MG, GSP |
 |
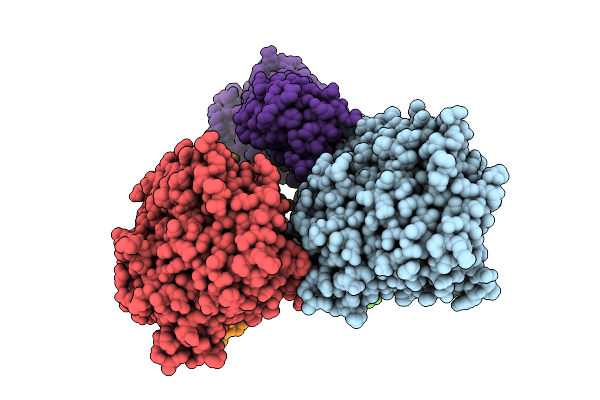
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Resolution:3.19 Å Release Date: 2025-11-12 Classification: STRUCTURAL PROTEIN Ligands: MG, GDP, EPB, GTP |
 |
Organism: Xenopus laevis, Bos taurus
Method: ELECTRON MICROSCOPY Resolution:2.89 Å Release Date: 2025-11-05 Classification: CELL CYCLE Ligands: G2P, MG, GTP |
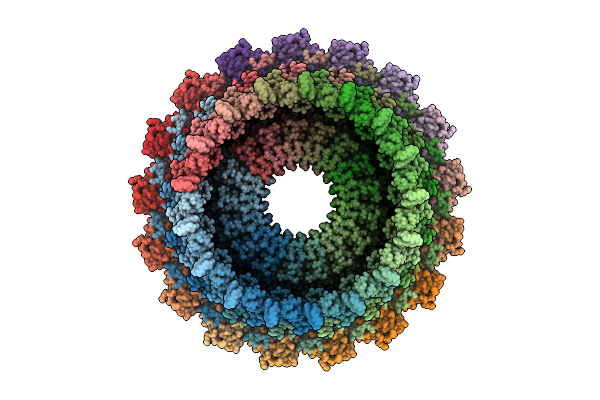 |
Organism: Escherichia coli o127:h6 str. e2348/69
Method: ELECTRON MICROSCOPY Resolution:2.56 Å Release Date: 2025-11-05 Classification: PROTEIN TRANSPORT |
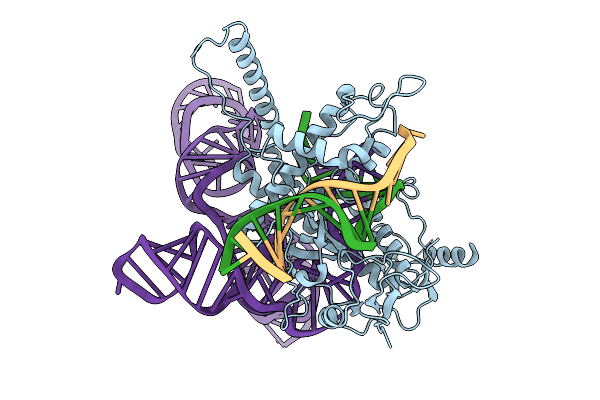 |
Structure Of Latranc Complex Bound To 27Nt Complementary Dna Substrate, Conformation 1
Organism: Lawsonibacter sp.
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: IMMUNE SYSTEM/DNA/RNA |

