Search Count: 705
 |
Cryo-Em Structure Of The Type Ii Secretion System Protein From Vibrio Cholerae
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PROTEIN TRANSPORT |
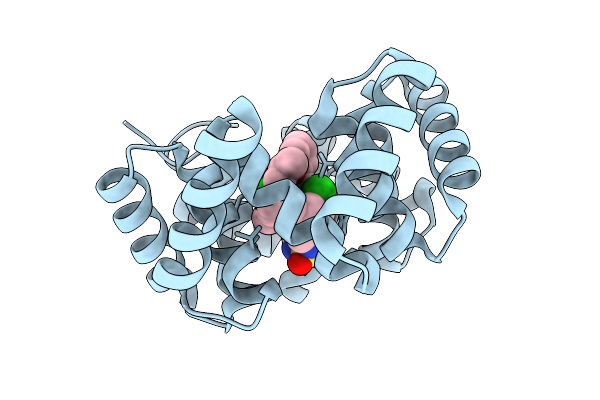 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: DE NOVO PROTEIN Ligands: A1IR7 |
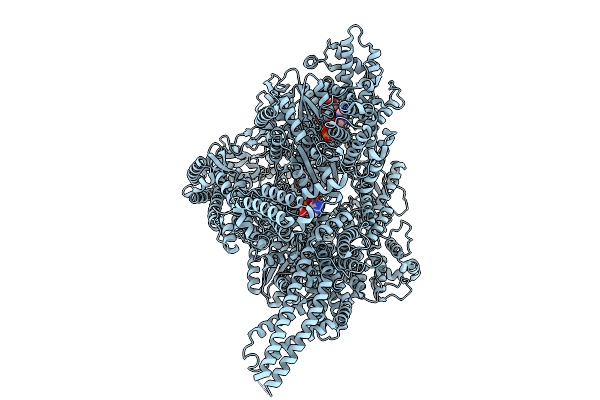 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MOTOR PROTEIN Ligands: ADP, ATP, ANP, MG |
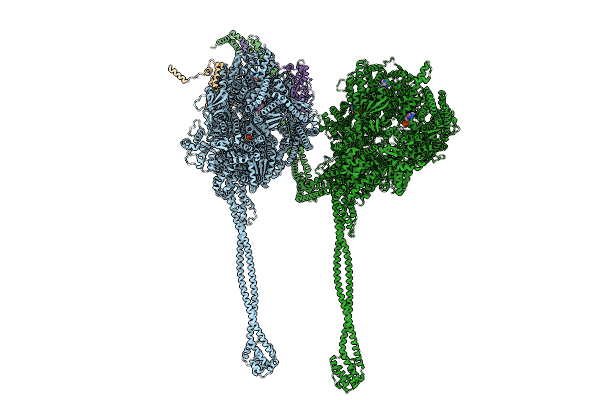 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MOTOR PROTEIN Ligands: ADP, ATP, MG, ANP |
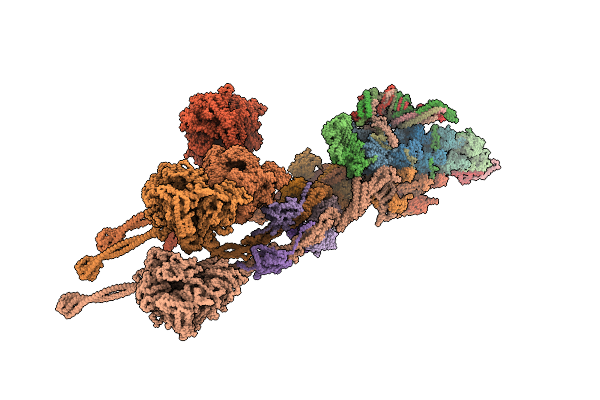 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MOTOR PROTEIN Ligands: ZN, MG |
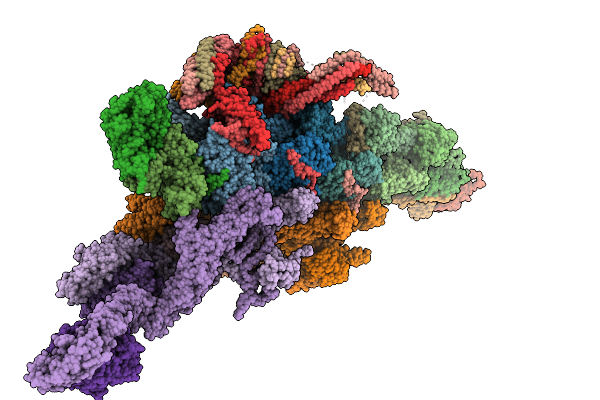 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MOTOR PROTEIN Ligands: ADP, ATP, ZN |
 |
Structure Of Dynactin, Dynein Tail With One Bicdr From Dynein-Dynactin-Bicdr On Microtubules
Organism: Mus musculus, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MOTOR PROTEIN Ligands: ADP, ATP, ZN |
 |
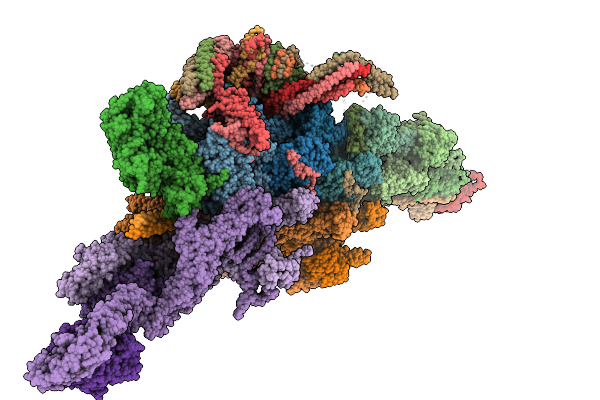
Structure Of Dynactin, Dynein Tail With Two Bicdr From Dynein-Dynactin-Bicdr On Microtubules
Organism: Mus musculus, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MOTOR PROTEIN Ligands: ADP, ATP, ZN |
 |
Structure Of Dynactin, Dynein Tail With Trak2 From Dynein-Dynactin-Trak2 On Microtubules
Organism: Mus musculus, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: MOTOR PROTEIN Ligands: ADP, ATP, ZN |
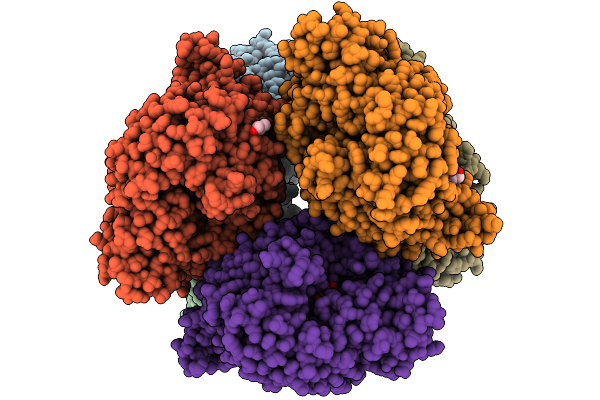 |
Structural Basis Of The Bifunctionality Of M. Salinexigens Zyf650T Glucosylglycerol Phosphorylase In Glucosylglycerol Catabolism
Organism: Marinobacter salinexigens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: STRUCTURAL PROTEIN Ligands: GLC, GOL, NA, G1P |
 |
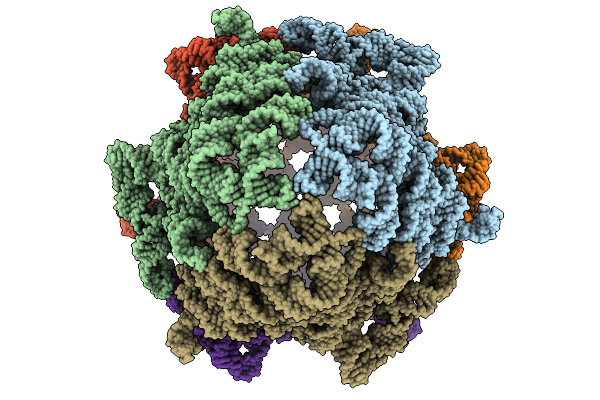
Structural Basis Of The Bifunctionality Of M. Salinexigens Zyf650T Glucosylglycerol Phosphorylase In Glucosylglycerol Catabolism
Organism: Marinobacter salinexigens
Method: X-RAY DIFFRACTION Release Date: 2025-09-10 Classification: STRUCTURAL PROTEIN Ligands: GOL, GLC, PEG |
 |
Organism: Ligilactobacillus salivarius ucc118
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Ligilactobacillus salivarius ucc118
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Ligilactobacillus salivarius ucc118
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Streptococcus agalactiae 18rs21
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Streptococcus agalactiae 18rs21
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Streptococcus agalactiae 18rs21
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Streptococcus agalactiae 18rs21
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Uncultured caudovirales phage
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |
 |
Organism: Uncultured caudovirales phage
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: RNA |

