Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Saccharomyces cerevisiae s288c
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: GENE REGULATION |
 |
Organism: Streptococcus macedonicus
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Crimean-Congo Hemorrhagic Fever Virus Cap-Snatching Endonuclease
Organism: Orthonairovirus haemorrhagiae, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE |
 |
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE |
 |
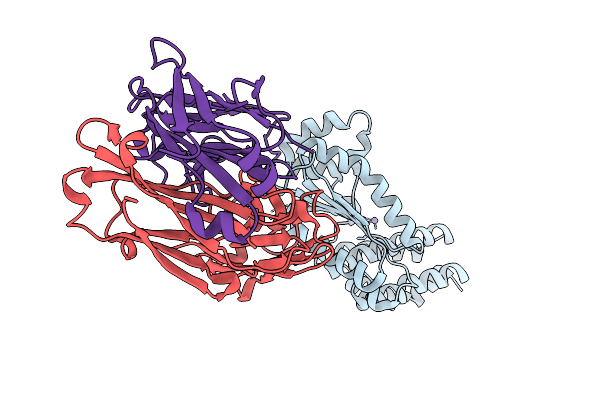
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With Manganese Ions
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN |
 |
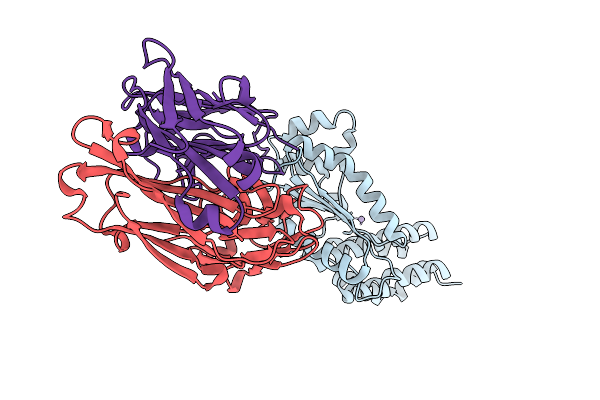
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease (E668A Mutant)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN |
 |
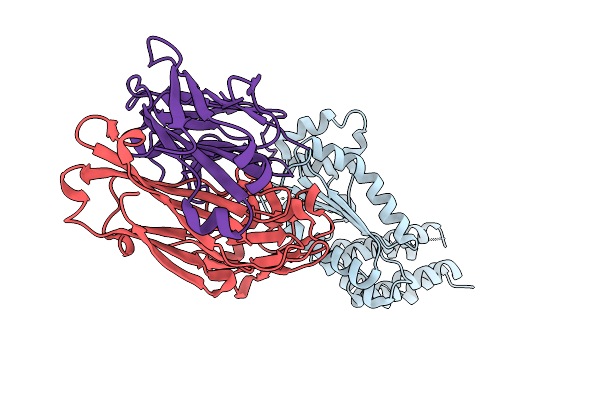
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease (E682A Mutant)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease (D719A Mutant)
Organism: Mus musculus, Kasokero virus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With 2,4-Dioxo-4-Phenylbutanoic Acid (Dpba)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN, XI7 |
 |
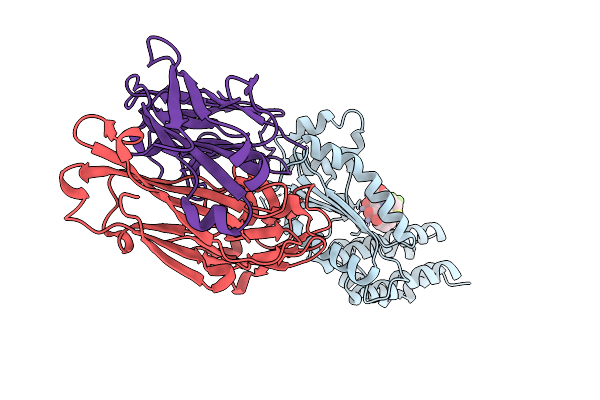
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With L-742,001
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN, 0N8 |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With Baloxavir Acid (Bxa)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN, E4Z |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN Ligands: ND0, SO4, TRS |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/HYDROLASE |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN Ligands: 9N8, SO4 |
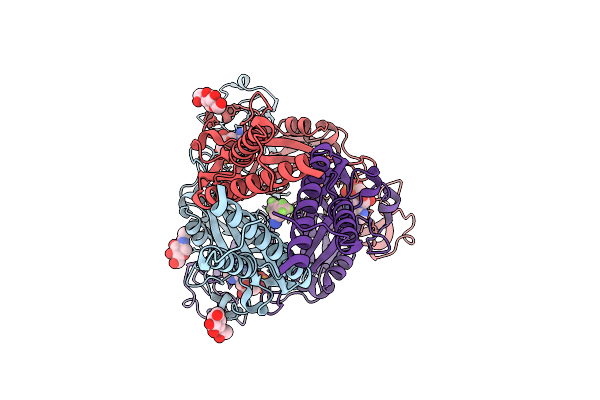 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: ATP, NAG, CA, MG, A1D8Z |
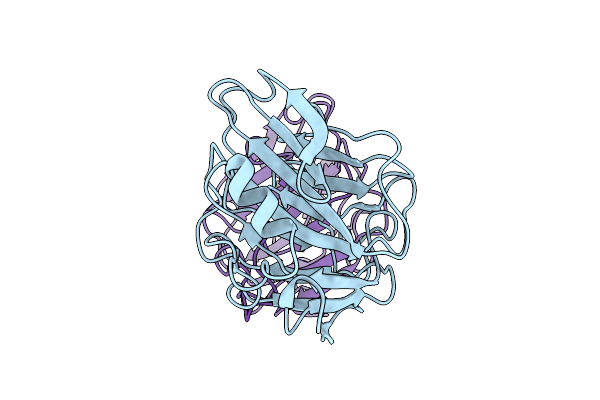 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
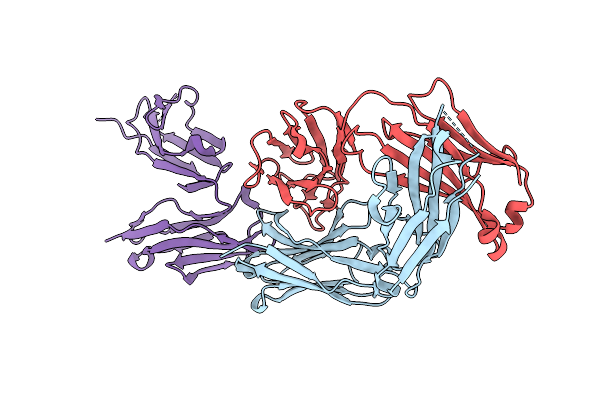 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
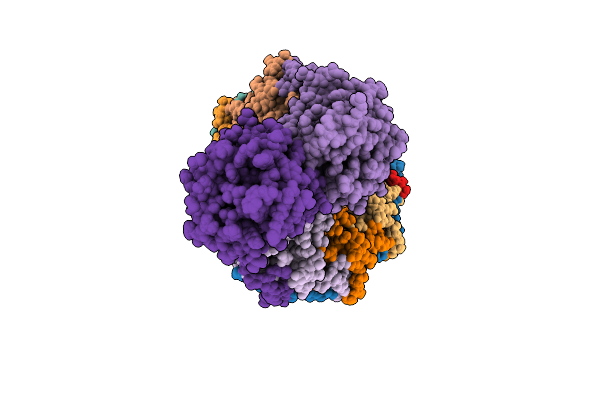 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: ANTIVIRAL PROTEIN/DNA/RNA Ligands: ATP, ZN, MG |