Search Count: 327
 |
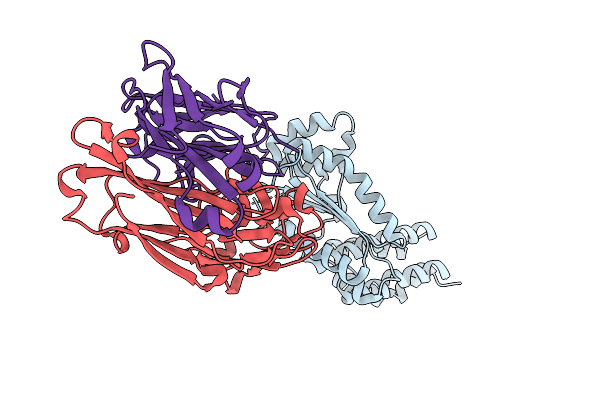
Crystal Structure Of Crimean-Congo Hemorrhagic Fever Virus Cap-Snatching Endonuclease
Organism: Orthonairovirus haemorrhagiae, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE |
 |
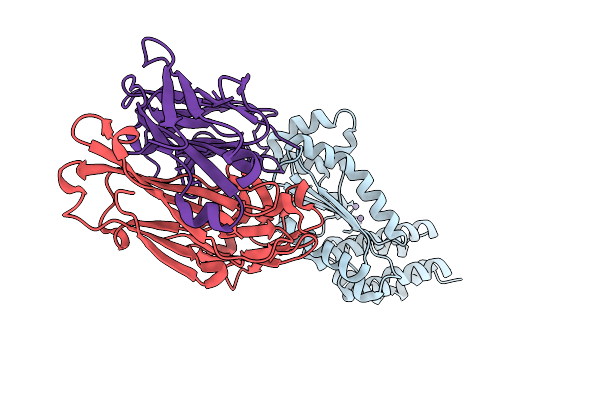
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE |
 |
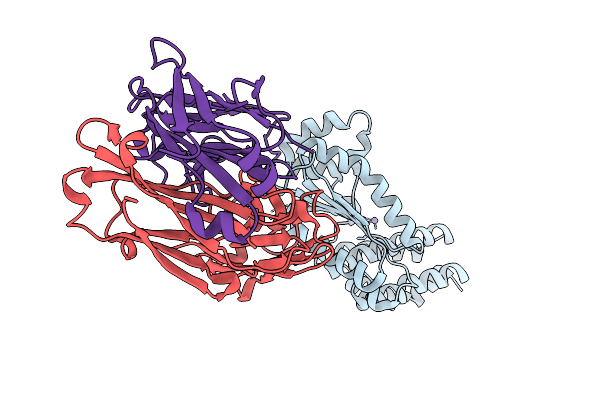
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With Manganese Ions
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN |
 |
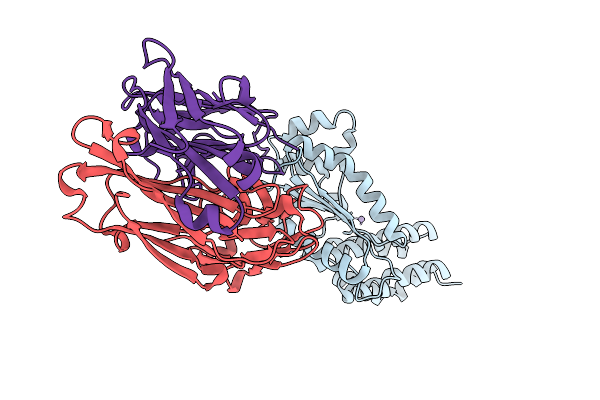
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease (E668A Mutant)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease (E682A Mutant)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease (D719A Mutant)
Organism: Mus musculus, Kasokero virus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With 2,4-Dioxo-4-Phenylbutanoic Acid (Dpba)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN, XI7 |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With L-742,001
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN, 0N8 |
 |
Crystal Structure Of Kasokero Virus Cap- Snatching Endonuclease In Complex With Baloxavir Acid (Bxa)
Organism: Kasokero virus, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: MN, E4Z |
 |
Single-Particle Cryo-Em Structure Of The First Variant Of Mobilized Colistin Resistance (Mcr-1) In Its Ligand-Bound State
Organism: Escherichia coli, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: KDL, PEE |
 |
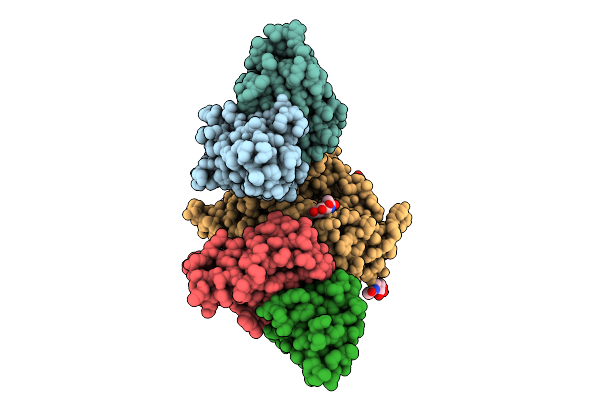
Organism: Henipavirus nipahense, Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
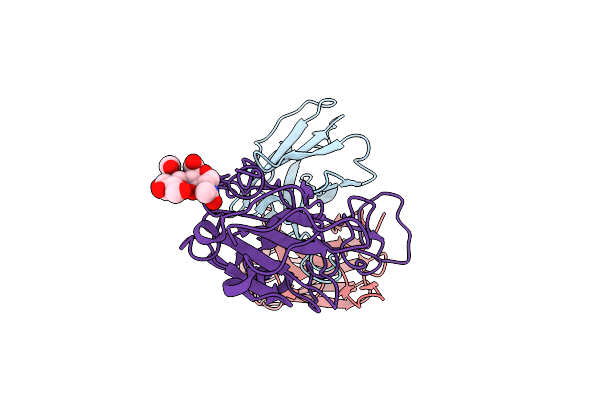
Organism: Henipavirus nipahense, Mesocricetus auratus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Cryo-Em Structure Of Sars-Cov-2 Prototype Spike Protein In Complex With H4 Fab (Local Refinement)
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
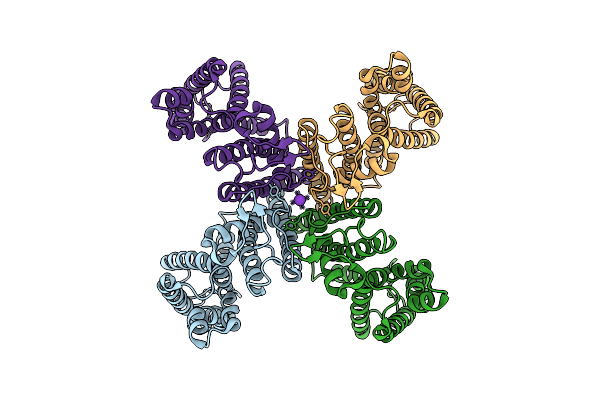 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PLANT PROTEIN Ligands: K |
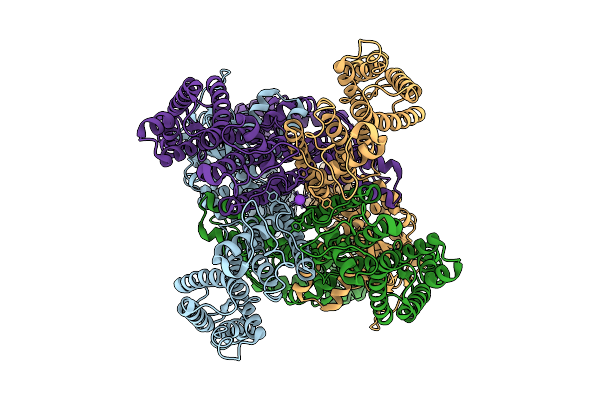 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PROTON TRANSPORT Ligands: K |
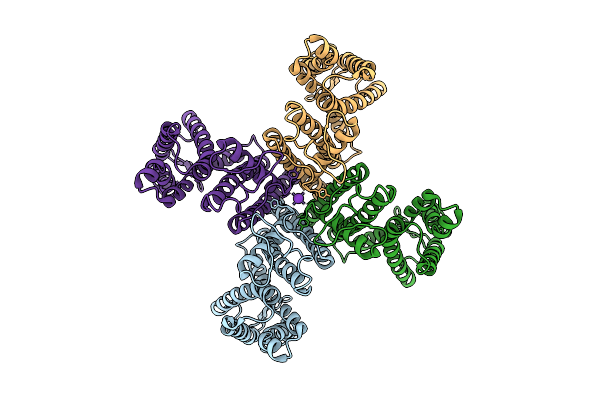 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PROTON TRANSPORT Ligands: K |
 |
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Inhibitor 1991
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: BIOSYNTHETIC PROTEIN Ligands: A1ECY, FMN, SF4, MG |
 |
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Inhibitor 1872
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2025-08-20 Classification: BIOSYNTHETIC PROTEIN Ligands: FMN, SF4, A1EEQ |
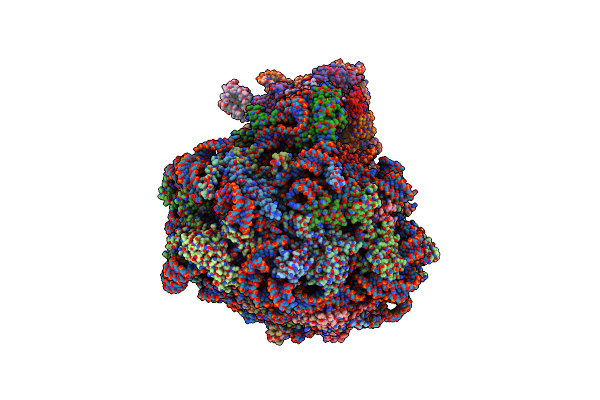 |
Single Particle Cryoem Structure Of The Pf80S Ribosome In Non-Rotated Pre State (Nrt A-P-E)
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: RIBOSOME |

