Search Count: 845
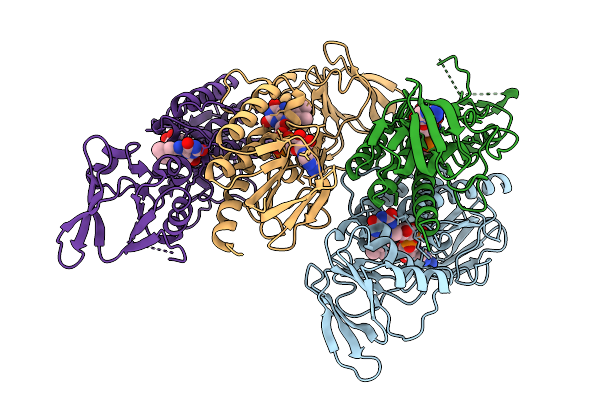 |
Crystal Structure Of Thioredoxin Reductase From Mycobacterium Tuberculosis.
Organism: Mycobacterium tuberculosis (strain cdc 1551 / oshkosh)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2026-02-04 Classification: OXIDOREDUCTASE Ligands: FAD |
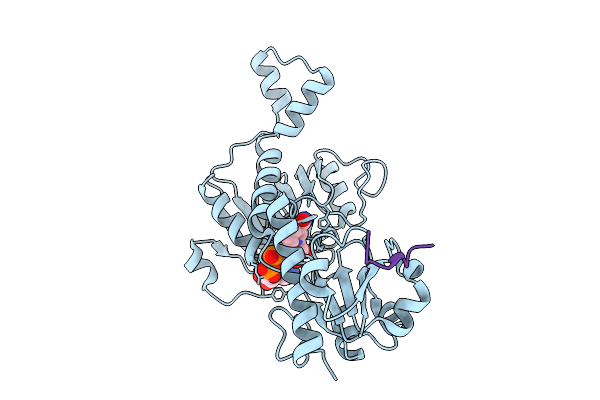 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2026-01-28 Classification: PROTEIN BINDING Ligands: NAD |
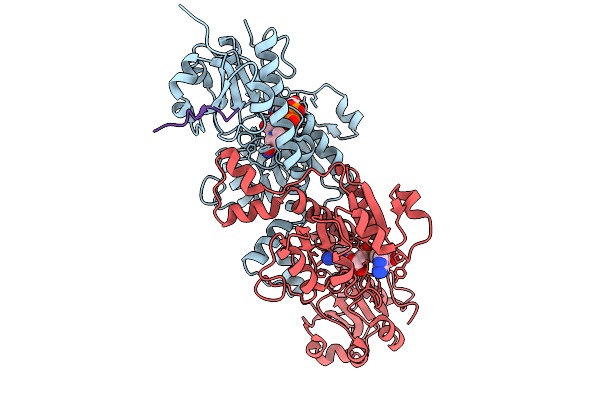 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2026-01-21 Classification: PROTEIN BINDING Ligands: NAD |
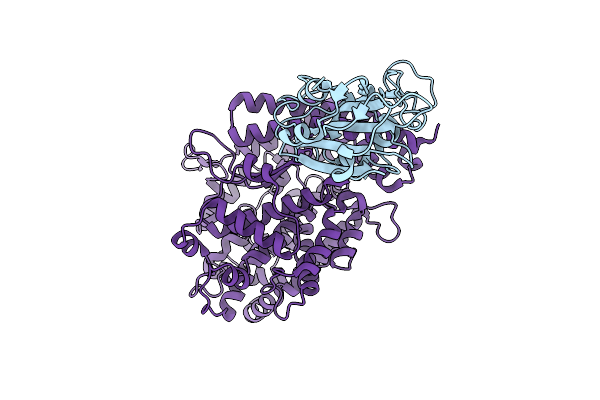 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/HYDROLASE |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/HYDROLASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.78 Å Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: NIO |
 |
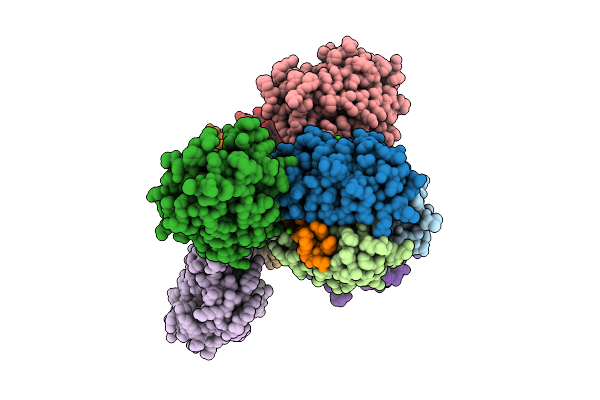
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2025-12-17 Classification: SIGNALING PROTEIN Ligands: CA, CAC, GOL, CL |
 |
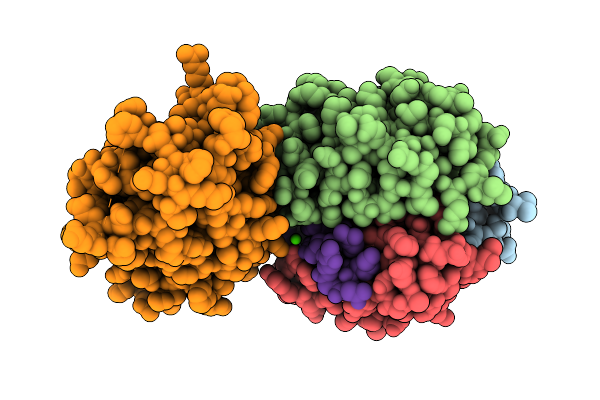
Organism: Mus musculus, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.48 Å Release Date: 2025-12-17 Classification: SIGNALING PROTEIN Ligands: CA |
 |
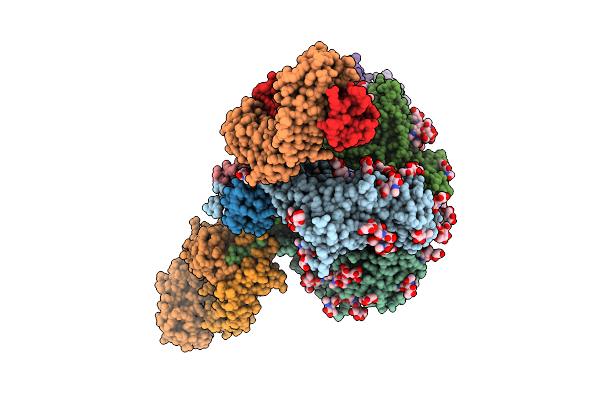
Organism: Mus musculus, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.34 Å Release Date: 2025-12-17 Classification: SIGNALING PROTEIN Ligands: CA |
 |
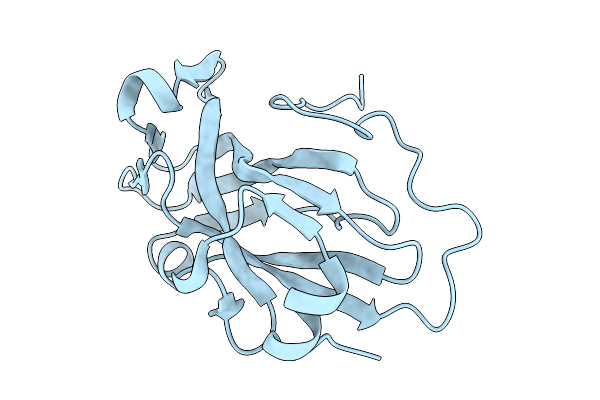
Cryo-Em Structure Of Hiv-1 Bg505Ds-Sosip.664 Env Trimer Bound To Dfph-A.01_10R59P_Lc Fab
Organism: Human immunodeficiency virus 1, Macaca
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-11-19 Classification: METAL BINDING PROTEIN |
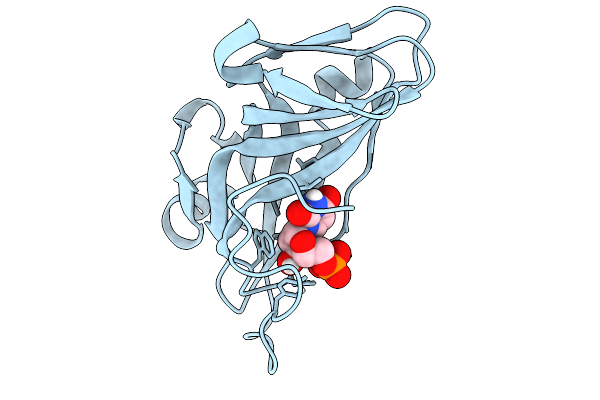 |
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-11-19 Classification: METAL BINDING PROTEIN Ligands: UMP |
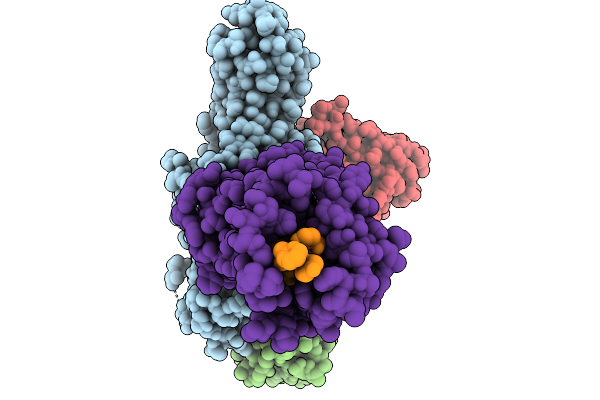 |
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 1)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
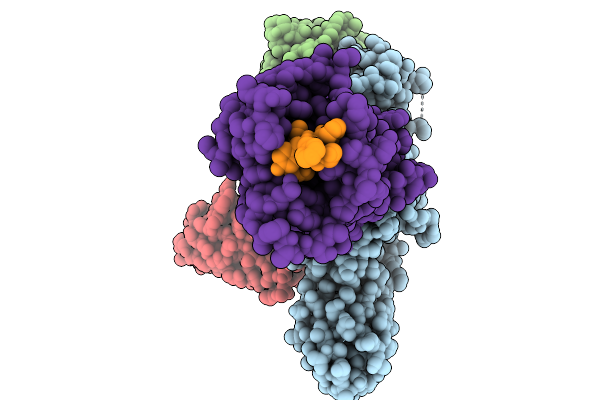 |
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 2)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
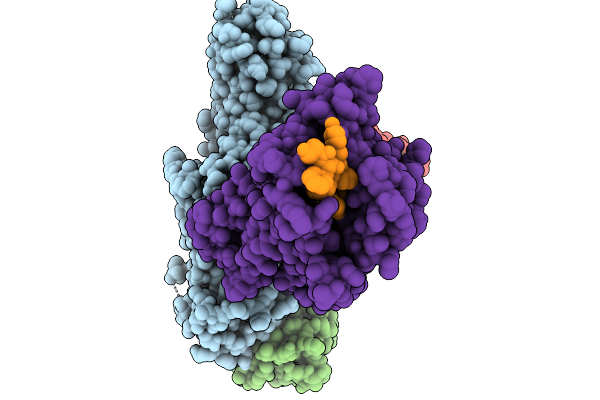 |
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 3)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
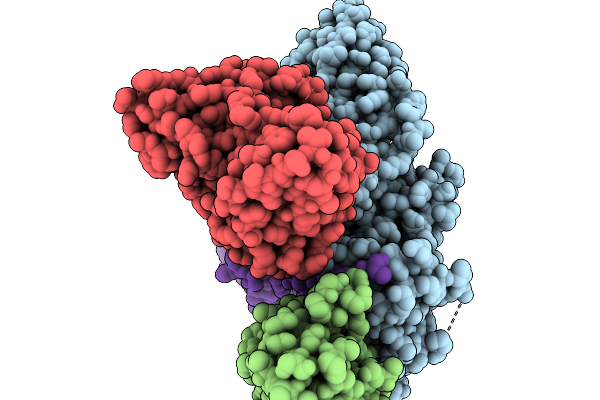 |
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 4)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
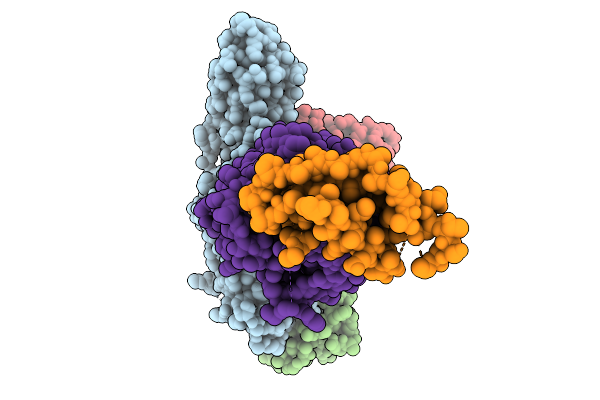 |
Composite Map Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 2
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Resolution:3.50 Å Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: Y01 |
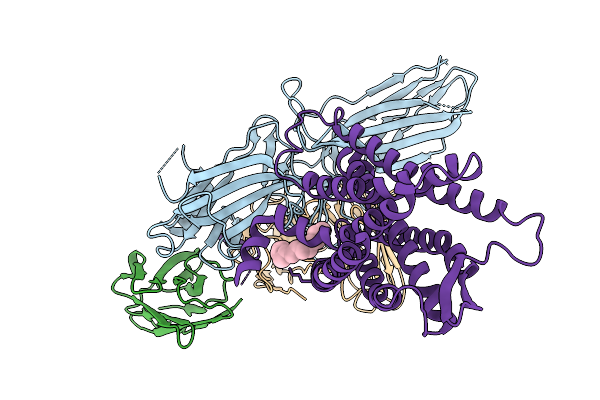 |
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Beta-Arrestin 1 In Ligand-Free State
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: PAM |
 |
The Crystal Structure Of Sars-Cov-2 Main Protease In Complex With Chebulagic Acid (Chla)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2025-11-05 Classification: VIRAL PROTEIN Ligands: A1D76, DMS |

