Search Count: 130
 |
Atomic Resolution Crystal Structure Of The Hexameric Antimicrobial Peptide Magainin-2
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2025-02-05 Classification: ANTIBIOTIC |
 |
The Structure Of The Human Tetrameric Ll-37 Peptide In A Channel Conformation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2021-09-08 Classification: ANTIMICROBIAL PROTEIN Ligands: CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.39 Å Release Date: 2021-07-14 Classification: CELL CYCLE Ligands: PT |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-29 Classification: STRUCTURAL PROTEIN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-07-29 Classification: STRUCTURAL PROTEIN |
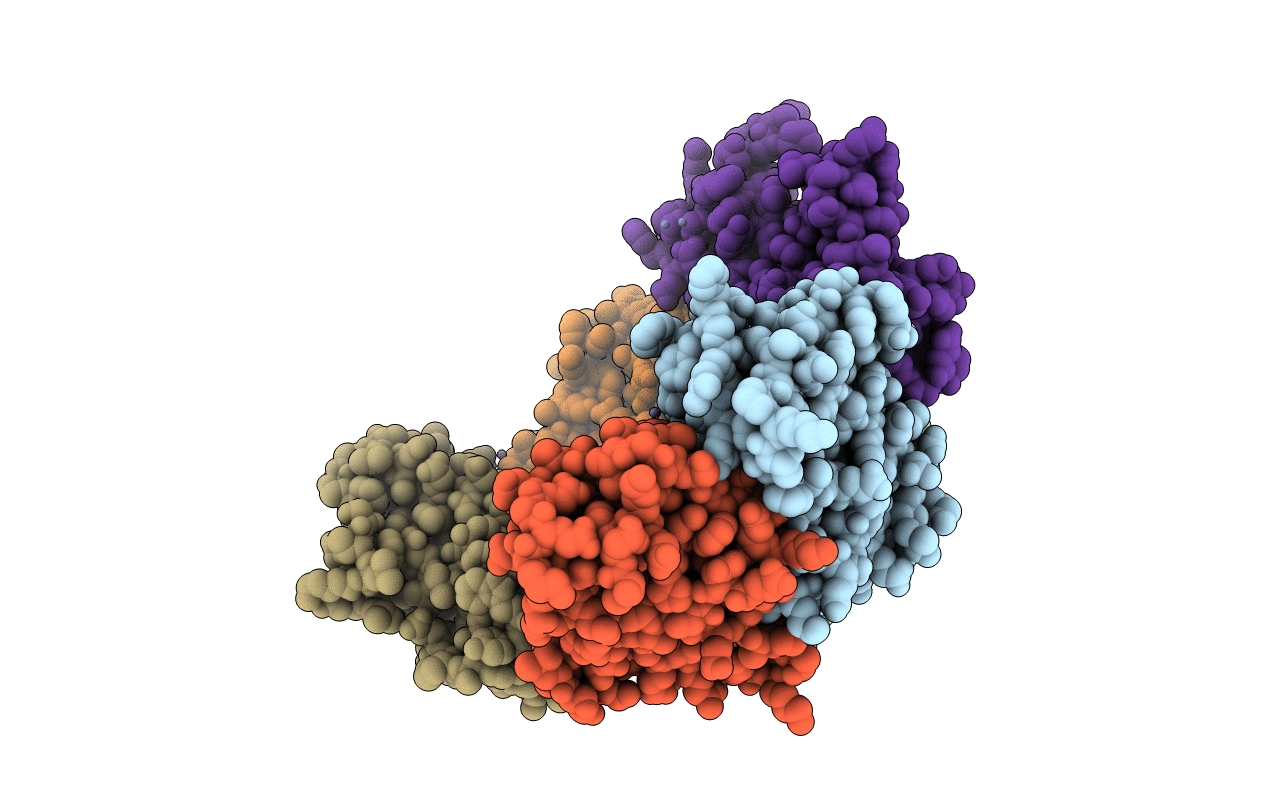 |
Structure Of Dps From Listeria Innocua Soaked With 10 Mm Zinc For 120 Minutes
Organism: Listeria innocua
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-09-25 Classification: METAL BINDING PROTEIN Ligands: ZN |
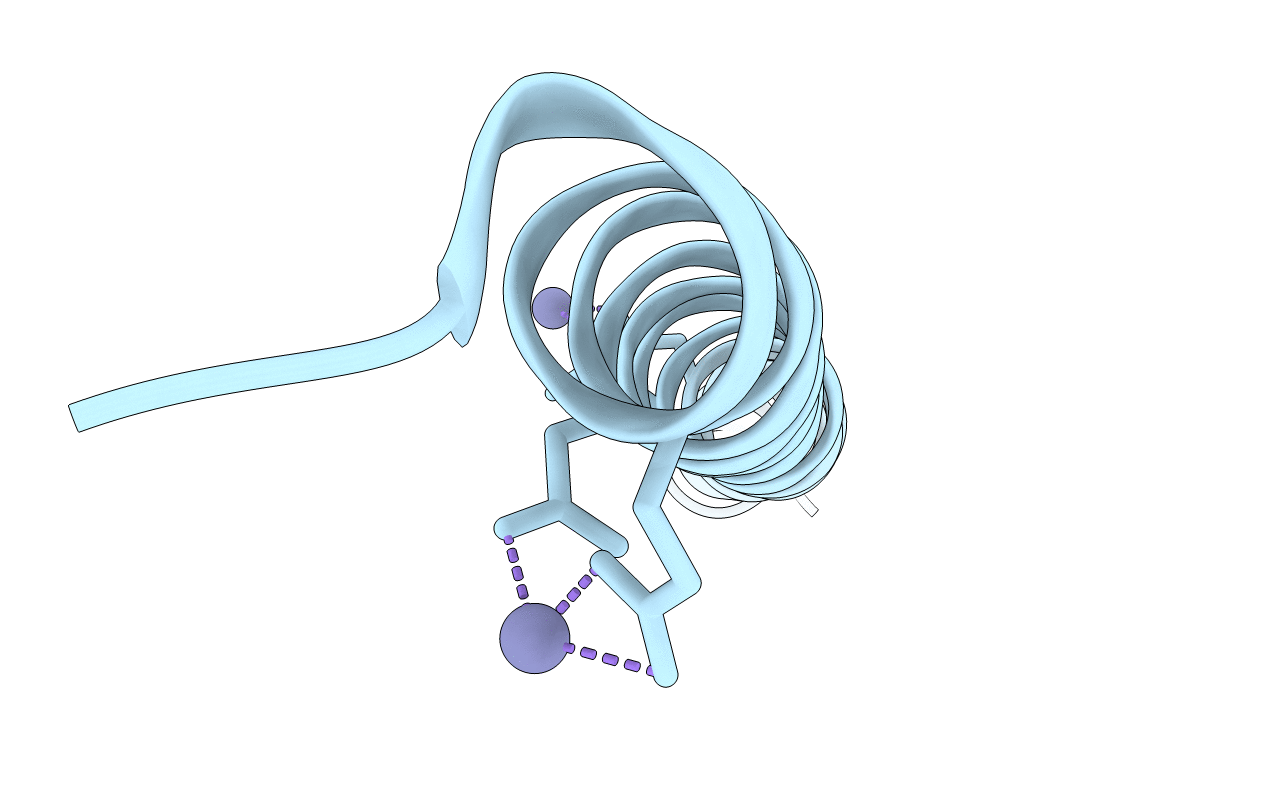 |
High Resolution Structure Of The Antimicrobial Peptide Dermcidin From Human
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2019-09-25 Classification: ANTIMICROBIAL PROTEIN Ligands: ZN |
 |
Organism: Listeria innocua
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-09-04 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
The Structure Of Dps From Listeria Innocua Soaked Before Soaking Experiments With Zn, Co And La
Organism: Listeria innocua serovar 6a (strain atcc baa-680 / clip 11262)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-09-04 Classification: METAL BINDING PROTEIN Ligands: LA |
 |
Organism: Listeria innocua clip11262
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-09-04 Classification: METAL BINDING PROTEIN Ligands: CO |
 |
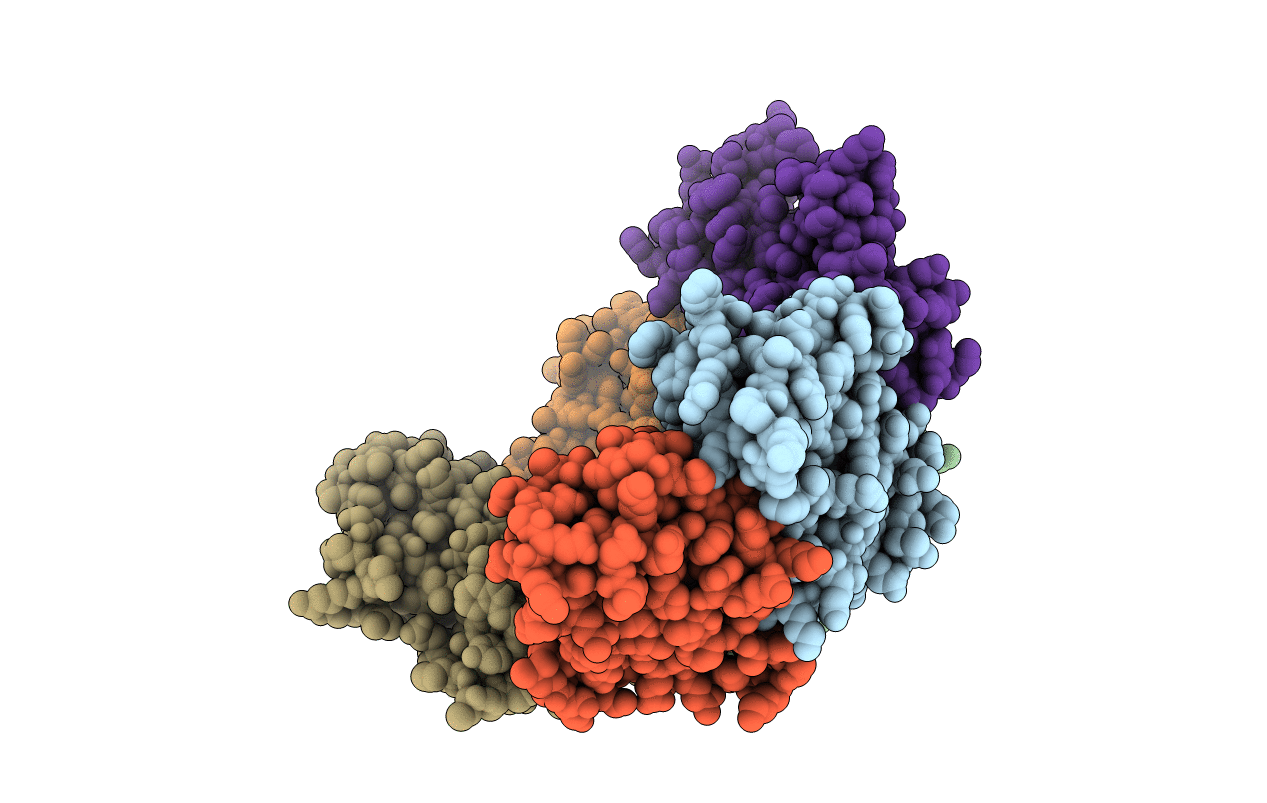
Crystal Structure Of The Ts2631 Endolysin From Thermus Scotoductus Phage With The Unique N-Terminal Moiety Responsible For Peptidoglycan Anchoring
Organism: Thermus phage 2631
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-01-30 Classification: ANTIMICROBIAL PROTEIN Ligands: ZN |
 |
The Apo Structure Of Dps From Listeria Innocua Before Soaking Experiments With Zn, Co And La
Organism: Listeria innocua
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2018-11-14 Classification: METAL BINDING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2018-01-24 Classification: ANTIMICROBIAL PROTEIN |
 |
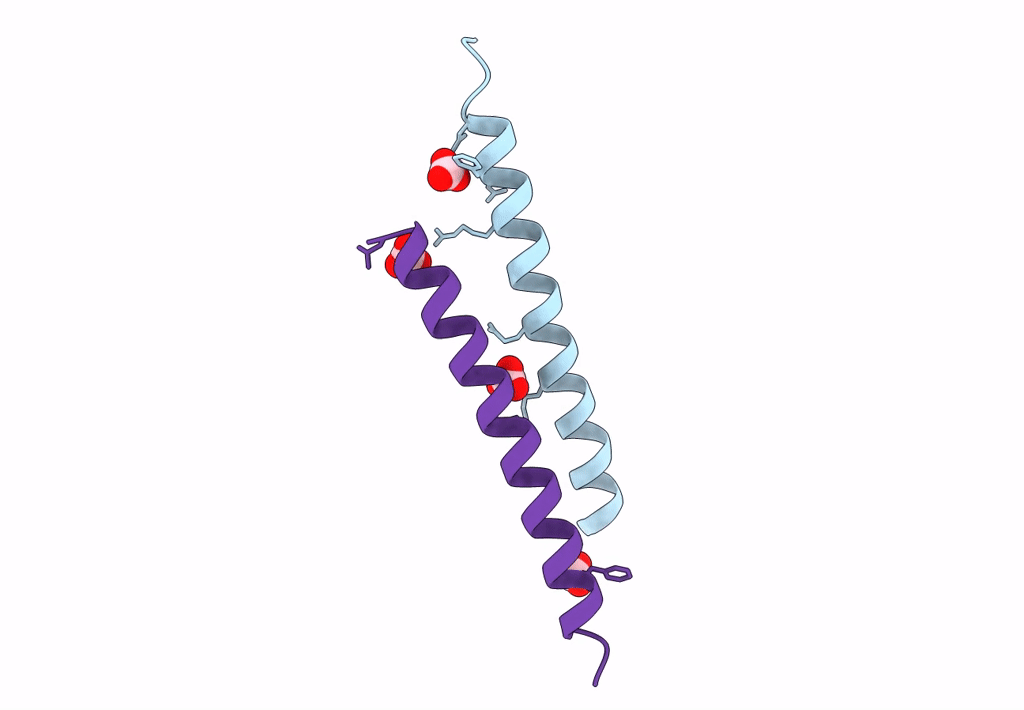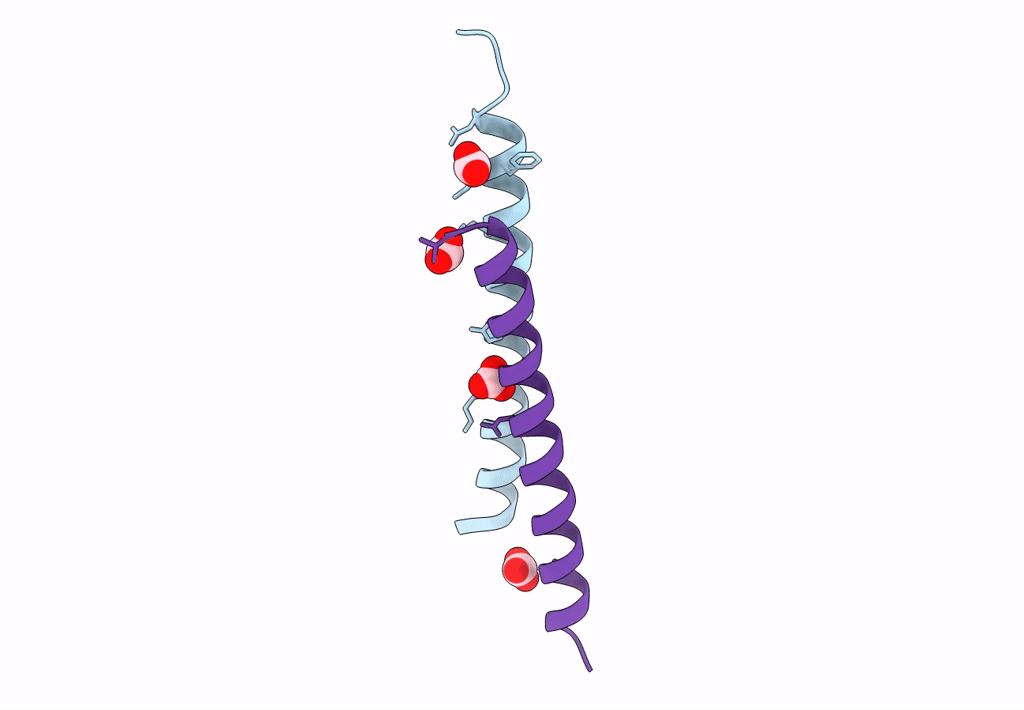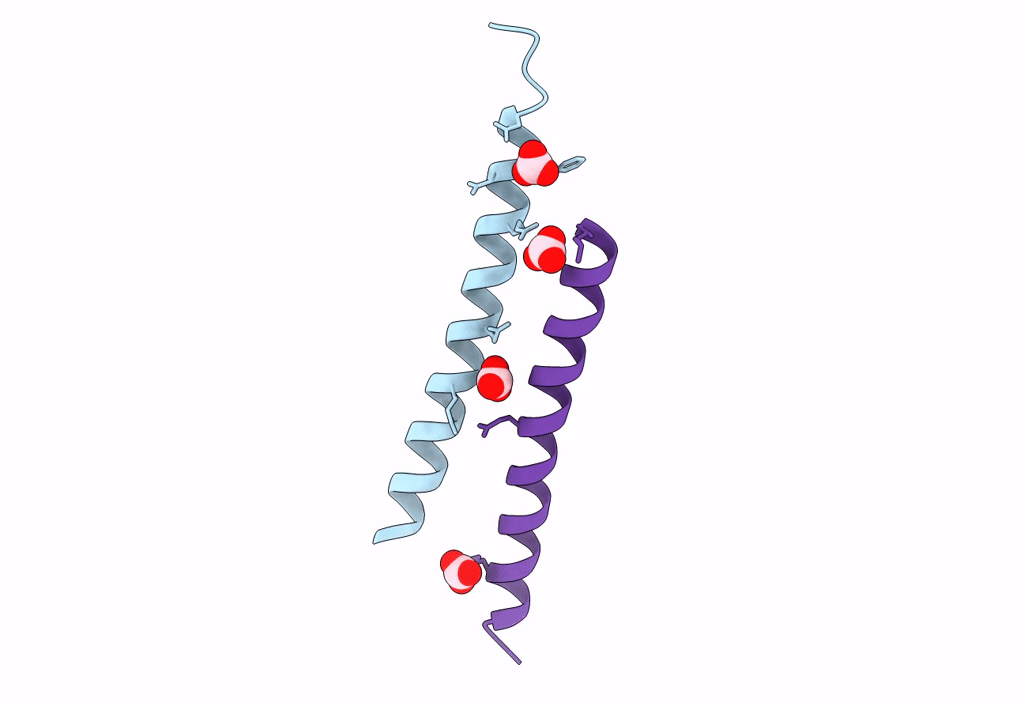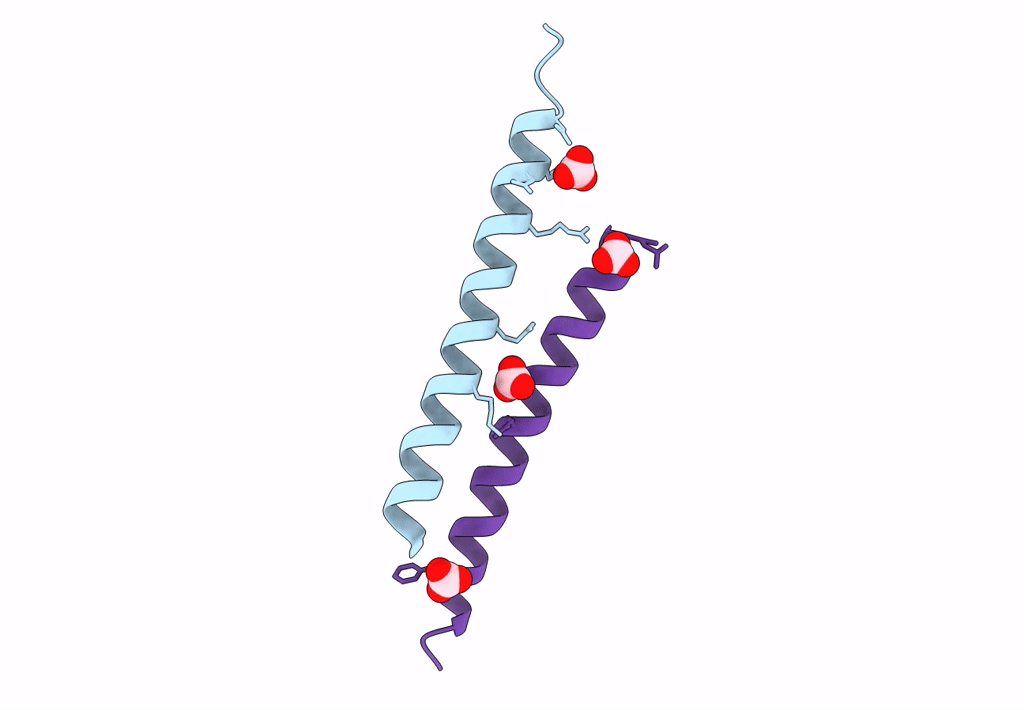
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-01-24 Classification: ANTIMICROBIAL PROTEIN Ligands: LDA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-01-24 Classification: ANTIMICROBIAL PROTEIN Ligands: CO3 |
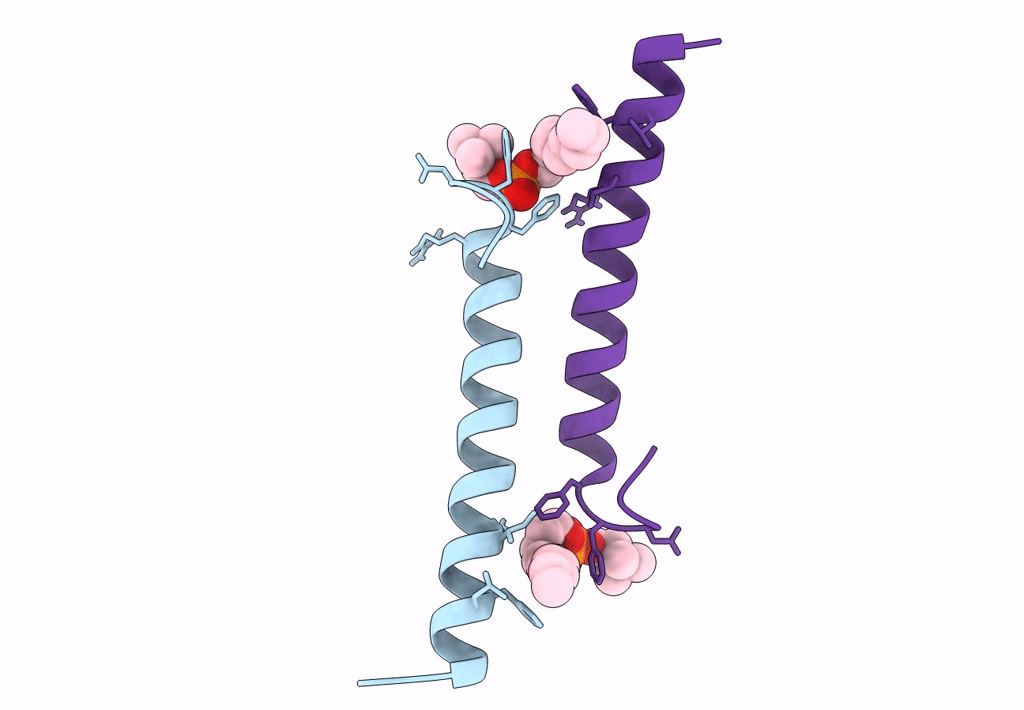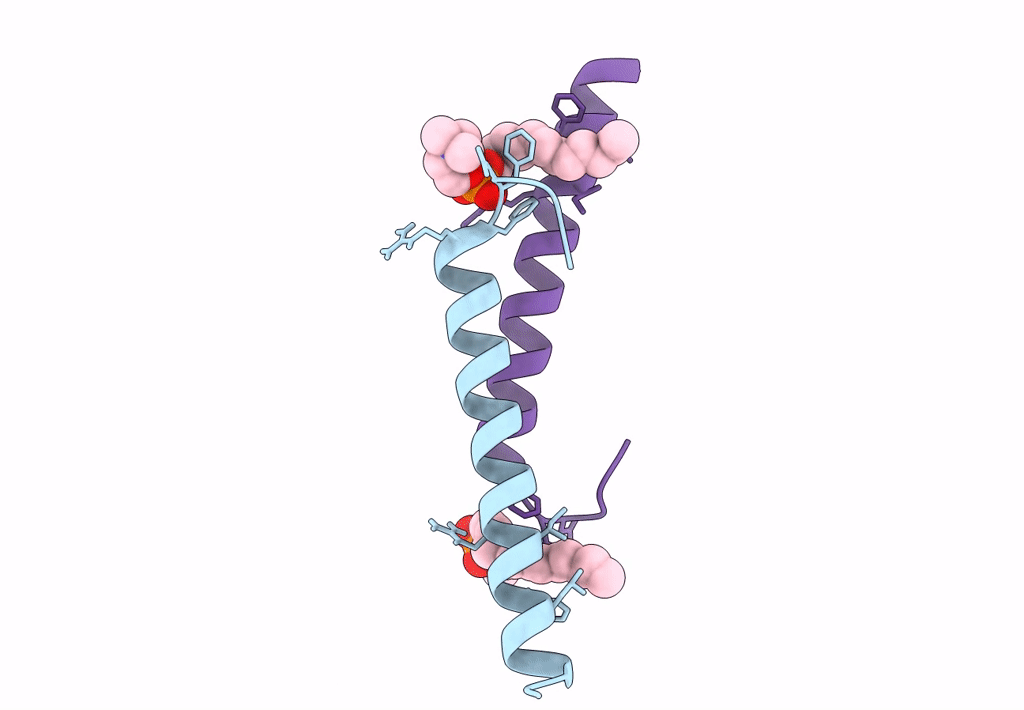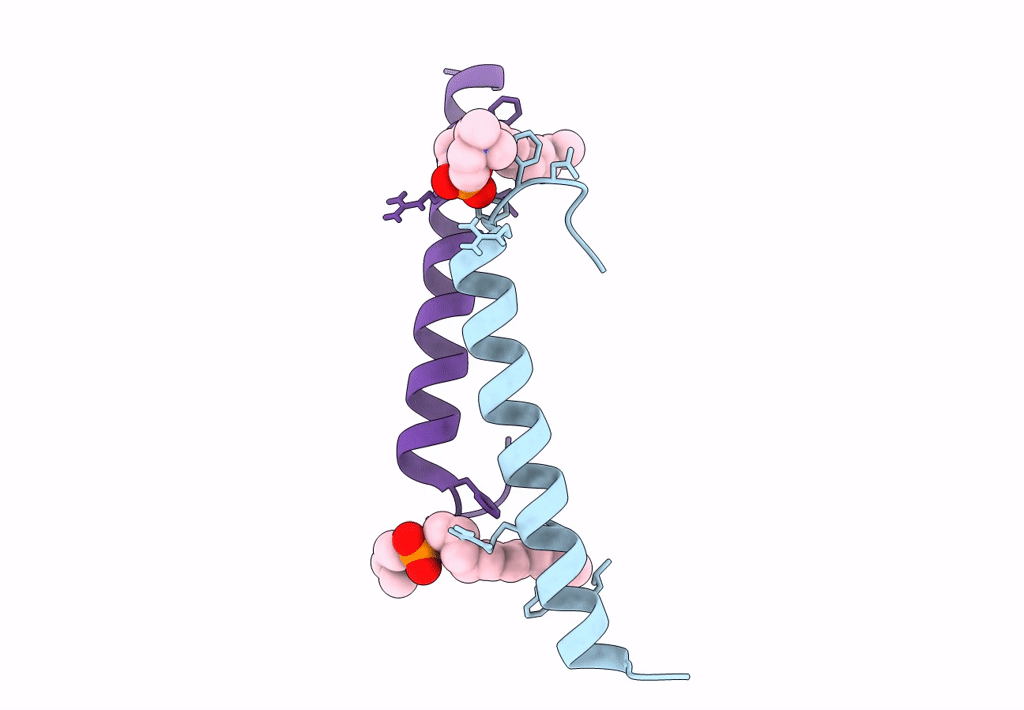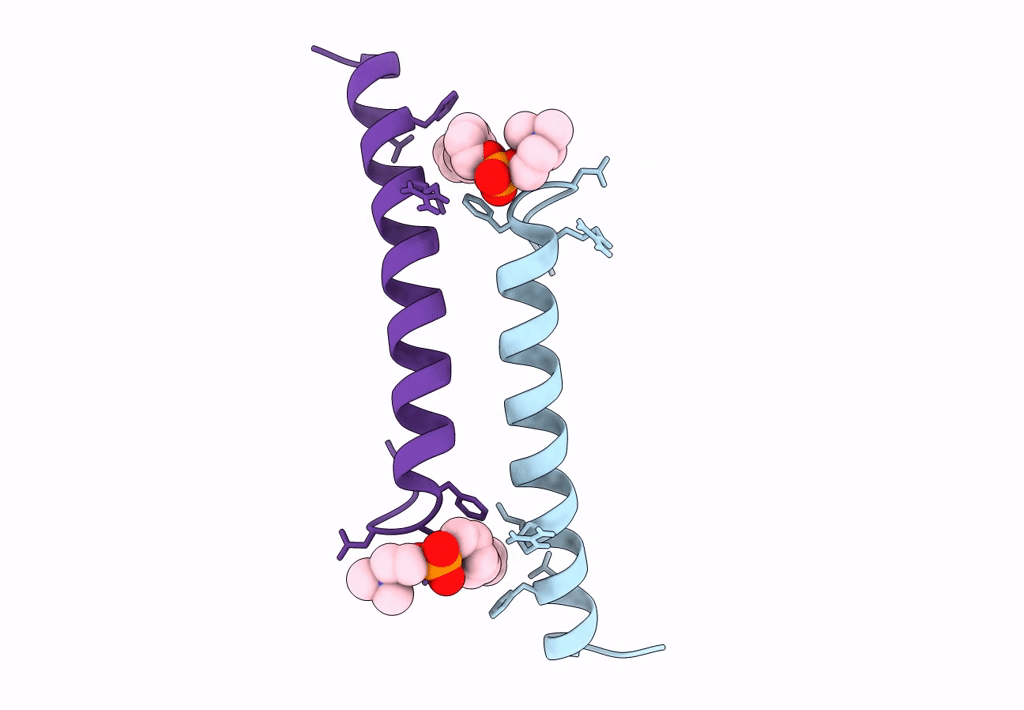 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2018-01-24 Classification: ANTIMICROBIAL PROTEIN Ligands: DPV |
 |
Organism: Salmonella enterica
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-06-24 Classification: SIGNALING PROTEIN |
 |
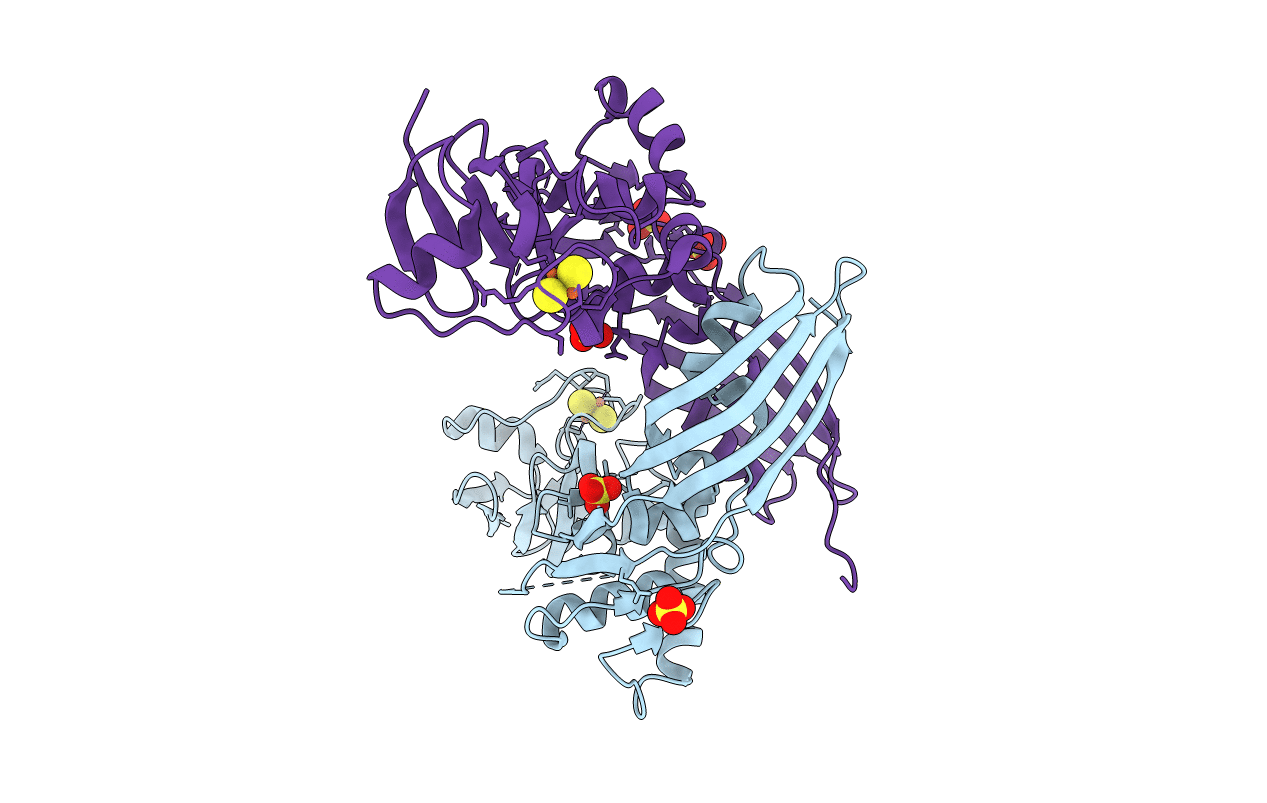
Organism: Pectobacterium carotovorum subsp. brasiliensis pbr1692
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2014-06-04 Classification: HYDROLASE Ligands: FES, SO4, GOL, MPD, CL |
 |
Organism: Pectobacterium carotovorum subsp. brasiliensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-06-04 Classification: HYDROLASE Ligands: FES, SO4, CL |
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2014-05-07 Classification: PROTEIN BINDING |

