Search Count: 75
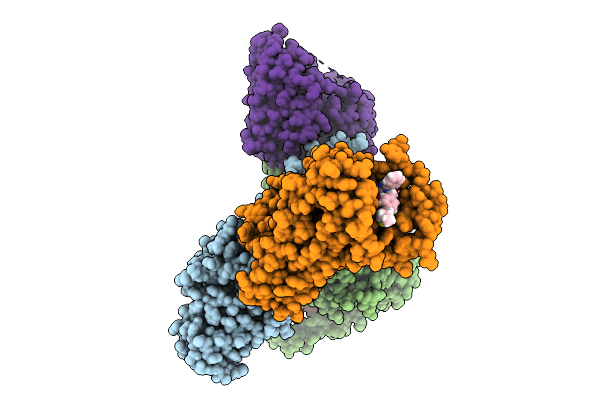 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: A1ECD |
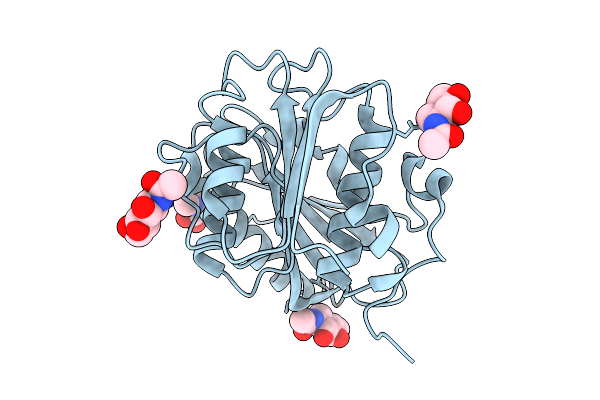 |
Crystal Structure Of Pichia Pastoris-Expressed Fast-Petase-N212A/K233C/S282C Variant
Organism: Piscinibacter sakaiensis
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE Ligands: NAG |
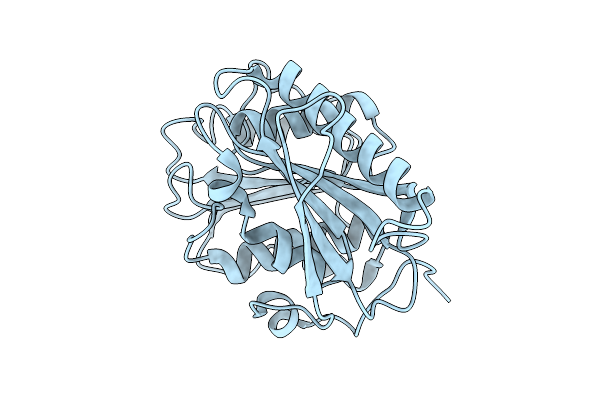 |
Organism: Piscinibacter sakaiensis
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE |
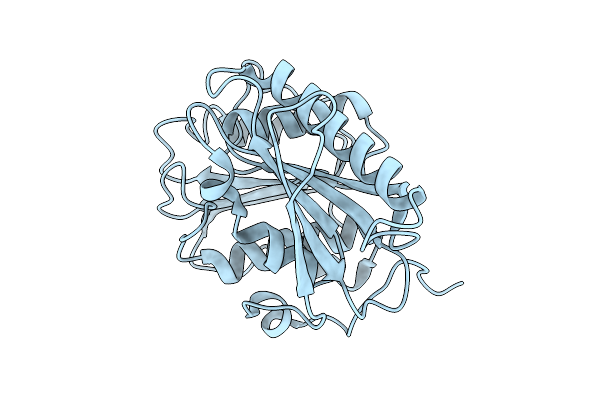 |
Organism: Piscinibacter sakaiensis
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE |
 |
Crystal Structure Of Fast-Acc-T140D/R132G/S160A Mutant In Complex With Mono(2-Hydroxyethyl) Terephthalic Acid
Organism: Piscinibacter sakaiensis
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE Ligands: C9C |
 |
Organism: Piscinibacter sakaiensis
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE |
 |
Inactive Mutant Of Fast-Acc-T140E/R132G In Complex With Mono(2-Hydroxyethyl) Terephthalic Acid
Organism: Piscinibacter sakaiensis
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: HYDROLASE Ligands: C9C |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: DNA BINDING PROTEIN/DNA Ligands: IHP, 2OP |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: MEMBRANE PROTEIN |
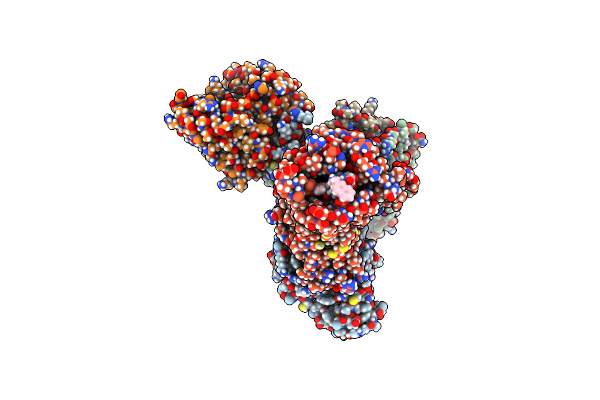 |
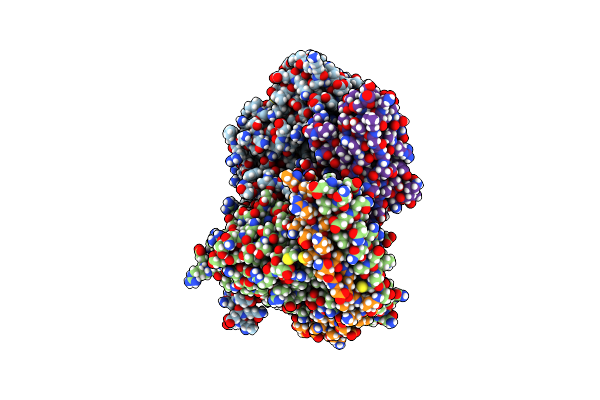
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: MEMBRANE PROTEIN |
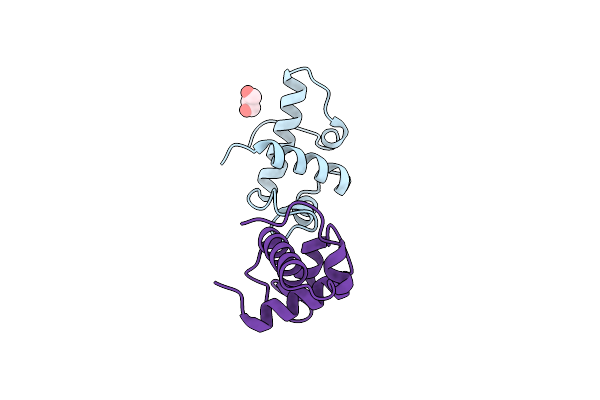 |
Crystal Structure Of The Myb Domain Of S.Pombe Tbf1 In The P222 Space Group
Organism: Schizosaccharomyces pombe 972h-
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2025-04-09 Classification: DNA BINDING PROTEIN Ligands: GOL |
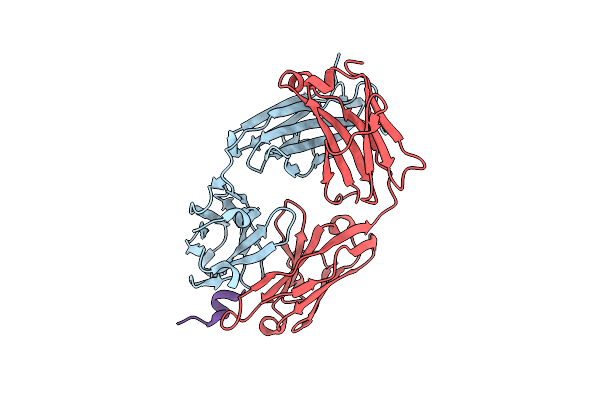 |
The Co-Crystal Structure Of The Fab Fragment Of Ab-1080 With Nav1.7 Vsdii Peptide
Organism: Oryctolagus cuniculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2024-10-23 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN Ligands: A1D8M |
 |
Cryo-Em Structure Of Gpr119-Gs Complex With Small Molecule Agonist Gsk-1292263
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: MEMBRANE PROTEIN Ligands: A1D8Y |
 |
Organism: Acinetobacter genomosp. 16bj
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: CELL ADHESION |
 |
Organism: Bacteria abnormis, Acinetobacter phage ap205
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: VIRUS/RNA |
 |
Organism: Acinetobacter phage ap205, Acinetobacter genomosp. 16bj
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: VIRUS |
 |
Organism: Acinetobacter phage ap205, Acinetobacter genomosp. 16bj
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: VIRUS |
 |
Organism: Acinetobacter phage ap205
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Acinetobacter phage ap205
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: VIRUS LIKE PARTICLE |

