Search Count: 117
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: TRANSPORT PROTEIN Ligands: X8M, NA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: CELL CYCLE Ligands: A1JBJ |
 |
Organism: Respiratory syncytial virus a2, Synthetic construct, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: HOH |
 |
Crystal Structure Of Nanobody Tnb04-1 With Antibody 1F11 Fab And Sars-Cov-2 Rbd
Organism: Vicugna pacos, Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
 |
Organism: Mus musculus, Rotavirus a
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: VIRUS |
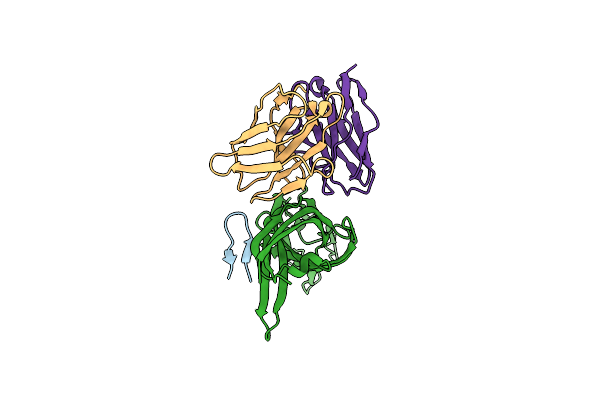 |
Cryo-Em Structure Of Simian Rotavirus Sa11 Vp4 In Complex With Nab 7H13-I54G Mutant (Left Side)
Organism: Mus musculus, Rotavirus a
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: VIRUS |
 |
Cryo-Em Structure Of Simian Rotavirus Sa11 Vp4 In Complex With Nab 7H13-I54G Mutant (Right Side)
Organism: Mus musculus, Rotavirus a
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: VIRUS |
 |
Organism: Bacillus sp. ps3
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: MEMBRANE PROTEIN Ligands: ANP, MG |
 |
Organism: Bacillus sp. ps3
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: HYDROLASE Ligands: ANP, MG, ADP, PO4 |
 |
Organism: Siraitia grosvenorii
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2024-09-04 Classification: TRANSFERASE Ligands: UD1 |
 |
Organism: Arabidopsis thaliana, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: STRUCTURAL PROTEIN Ligands: BEF, ADP |
 |
Organism: Arabidopsis thaliana, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: STRUCTURAL PROTEIN Ligands: ADP, BEF |
 |
Organism: Rhodopseudomonas palustris
Method: X-RAY DIFFRACTION Release Date: 2024-06-19 Classification: TRANSCRIPTION |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-02-28 Classification: MEMBRANE PROTEIN Ligands: PIO |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2024-02-28 Classification: MEMBRANE PROTEIN Ligands: PIO |
 |
Cryo-Em Structure Of Ebv Ghgl-Gp42 In Complex With Mabs 3E8 And 5E3 (Localized Refinement)
Organism: Human gammaherpesvirus 4, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: VIRAL PROTEIN |

