Search Count: 294
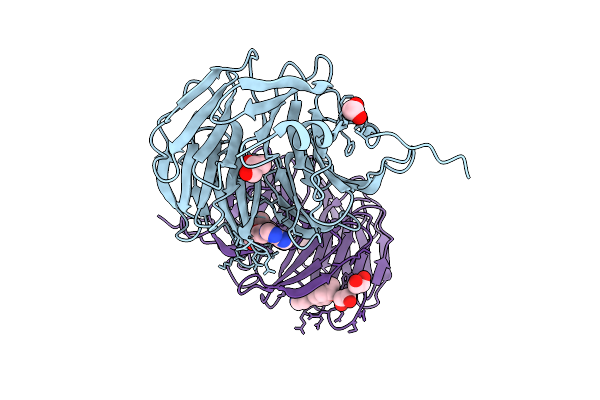 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: NUCLEAR PROTEIN Ligands: GOL, A1BI5, EDO |
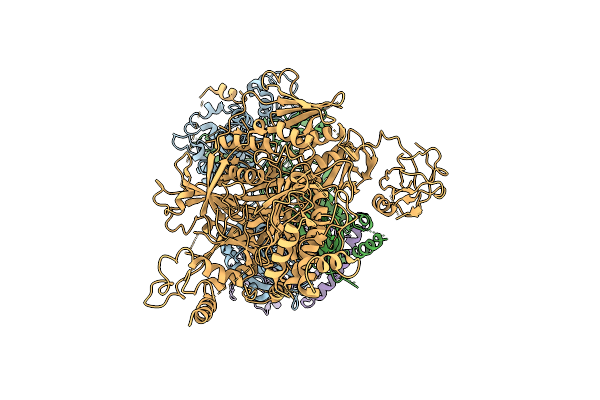 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: SIGNALING PROTEIN |
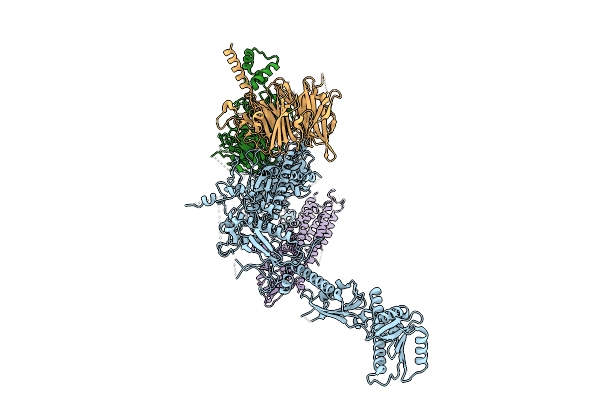 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: SIGNALING PROTEIN |
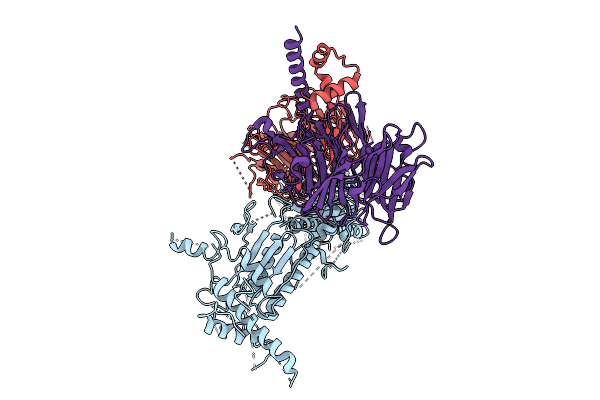 |
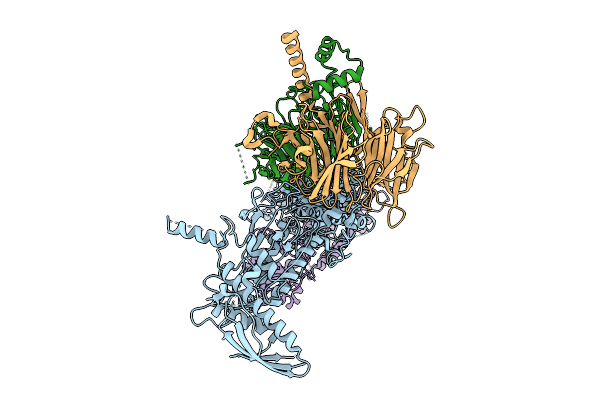
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: SIGNALING PROTEIN |
 |
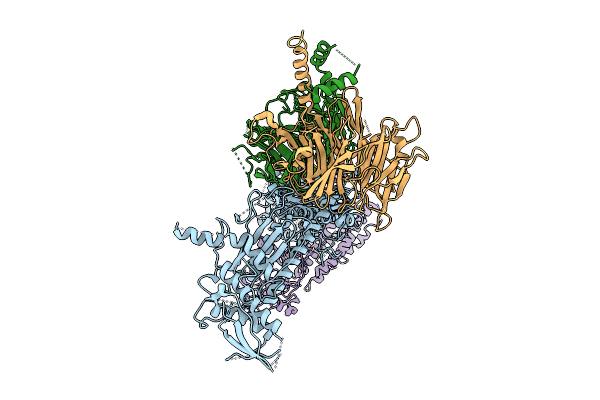
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: SIGNALING PROTEIN |
 |
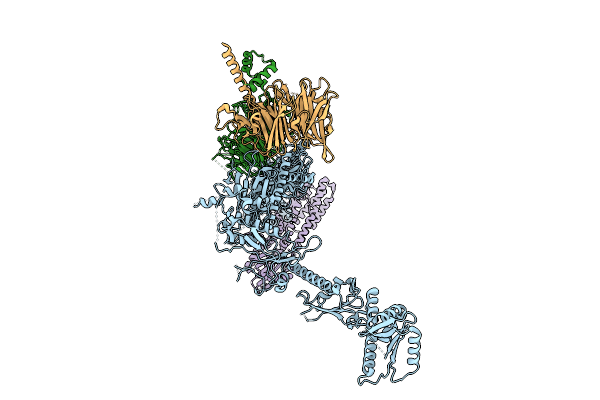
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: SIGNALING PROTEIN |
 |
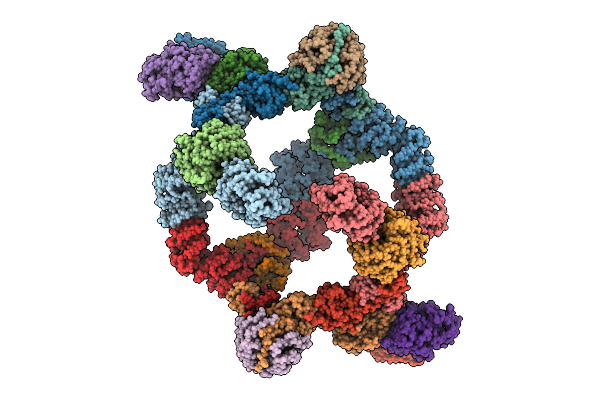
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: SIGNALING PROTEIN |
 |
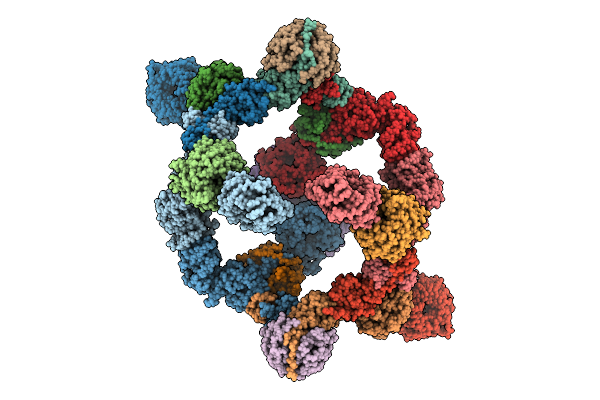
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
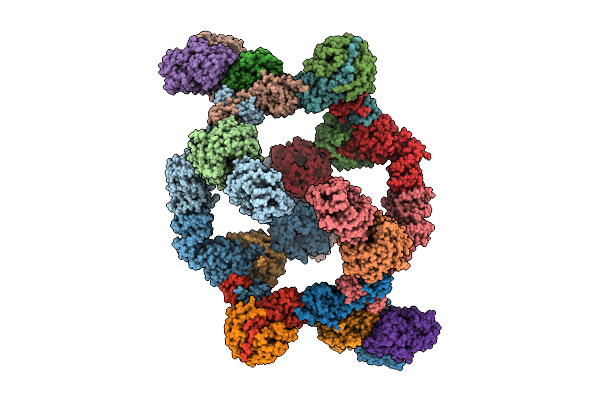
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: STRUCTURAL PROTEIN Ligands: ZN |
 |
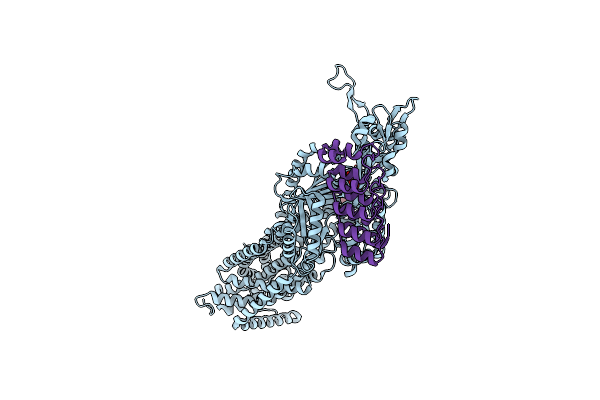
Organism: Alternaria alternata
Method: ELECTRON MICROSCOPY Release Date: 2025-06-18 Classification: TOXIN |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSPORT PROTEIN Ligands: MIY |
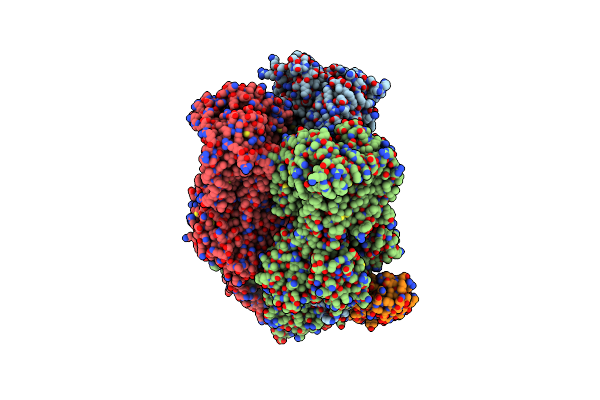 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSPORT PROTEIN |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: METAL BINDING PROTEIN |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
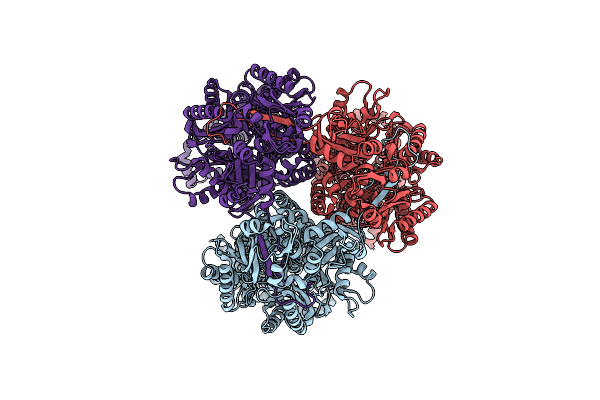 |
Single Particle Cryo-Em Structure Of The Multidrug Efflux Pump Oqxb From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
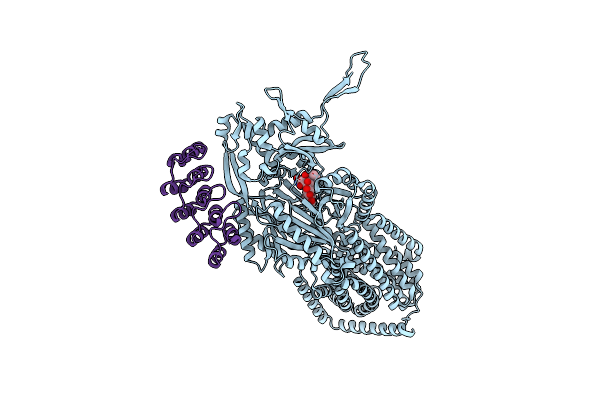 |
Organism: Synthetic construct, Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN Ligands: MIY |
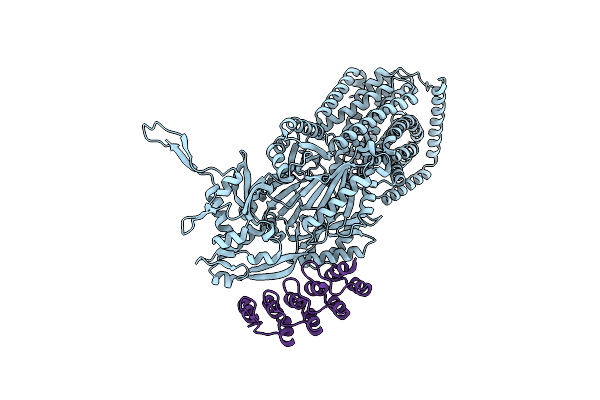 |
Organism: Synthetic construct, Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |

