Search Count: 18
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-11-02 Classification: CELL CYCLE Ligands: GDP, MG, NCO, MPD, CAC |
 |
Crystal Structure Of A Truncated Lsd1:Corest In The Presence Of An Lsd1-Nt Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-06-30 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2021-02-17 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: NAG |
 |
Crystal Structure Of The Swirm Domain Of Human Histone Lysine-Specific Demethylase Lsd1
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2019-07-10 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
The Molecular Mechanisms By Which Ns1 Of The 1918 Spanish Influenza A Virus Hijack Host Protein-Protein Interactions
Organism: Homo sapiens, Orthomyxoviridae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-08-08 Classification: protein binding/viral protein |
 |
Organism: Francisella tularensis subsp. novicida (strain u112)
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2017-08-16 Classification: TRANSCRIPTION |
 |
The Molecular Mechanisms By Which Ns1 Of The 1918 Spanish Influenza A Virus Hijack Host Protein-Protein Interactions
Organism: Homo sapiens, Influenza a virus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2017-08-09 Classification: VIRAL PROTEIN |
 |
Structure, Thermodynamics, And The Role Of Conformational Dynamics In The Interactions Between The N-Terminal Sh3 Domain Of Crkii And Proline-Rich Motifs In Cabl
Organism: Mus musculus, Endothia gyrosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-06-29 Classification: SIGNALING PROTEIN Ligands: PEG, NA, P4G |
 |
Crystal Structure Of Ev71 3Cpro In Complex With A Potent And Selective Inhibitor
Organism: Enterovirus a71
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2015-11-25 Classification: HYDROLASE Ligands: E22 |
 |
 |
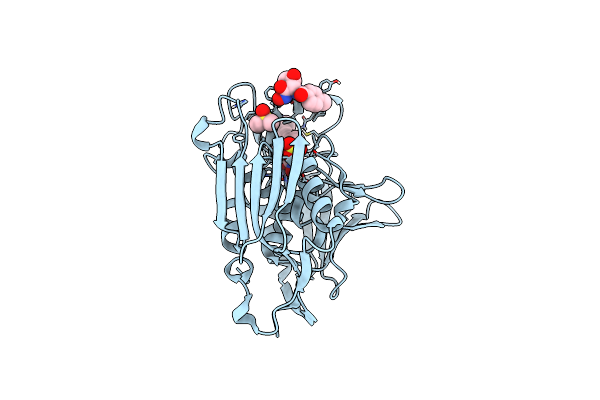
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2013-07-24 Classification: Transferase/transferase activator Ligands: GLC, 926 |
 |
 |
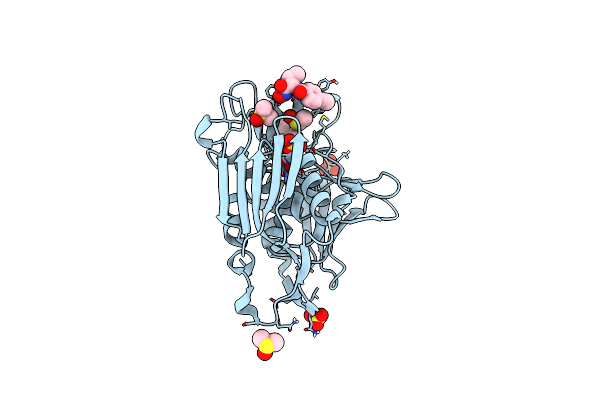
Organism: Escherichia coli ihe3034
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-01-19 Classification: HYDROLASE/ANTIBIOTIC Ligands: ZN, ZH2, UKW, SO4, DMS |
 |
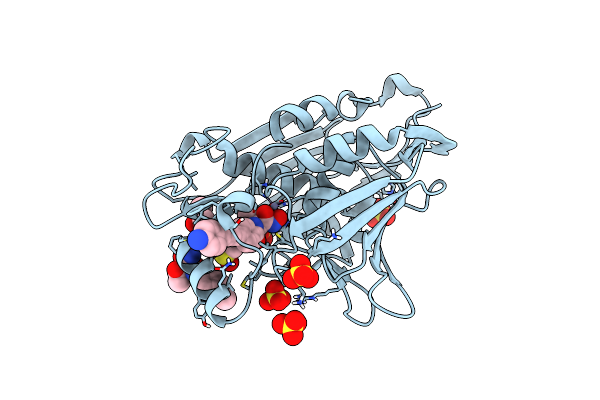
Organism: Escherichia coli ihe3034
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-01-19 Classification: HYDROLASE/ANTIBIOTIC Ligands: ZN, ZH4, UKW, DMS, SO4 |
 |
Organism: Escherichia coli ihe3034
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-01-19 Classification: HYDROLASE/ANTIBIOTIC Ligands: ZN, L53, UKW, DMS, SO4 |
 |
Organism: Aquifex aeolicus
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2011-01-05 Classification: HYDROLASE Ligands: ZN, 3P3, PO4 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2011-01-05 Classification: HYDROLASE Ligands: ZN, 3P3, NA, NO3 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2011-01-05 Classification: HYDROLASE Ligands: ZN, 3P3, SO4, DMS, UKW |

