Search Count: 20
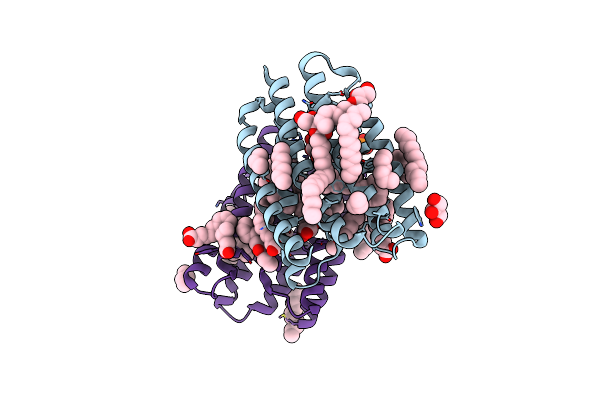 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The Ground State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
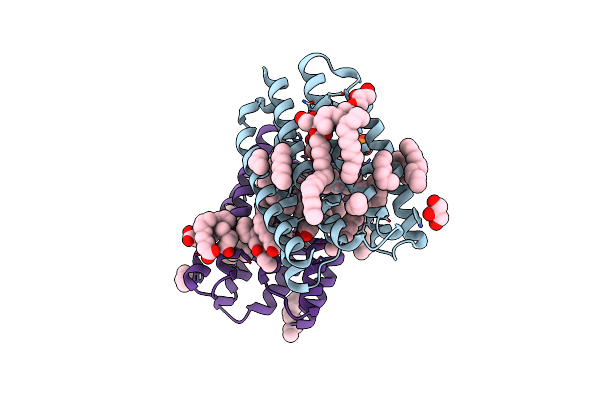 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The P596 State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLC, LFA, GOL, RET, PO4 |
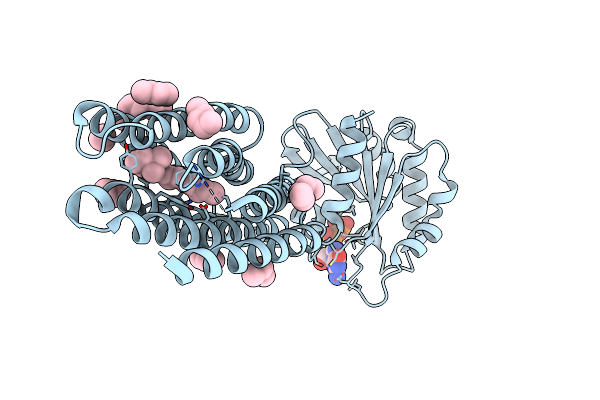 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; Ground State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, RET |
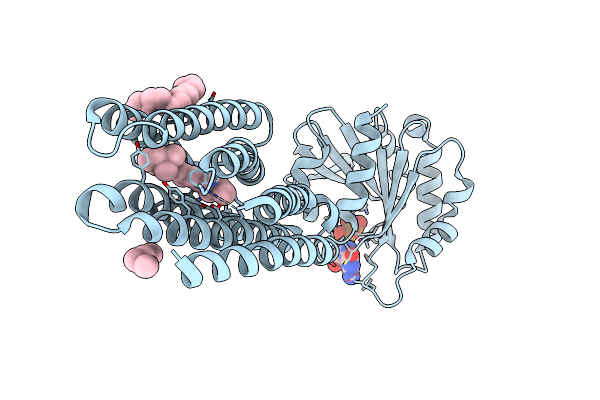 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) - Human Gtpase Arf1 (L8K,Q71L) Chimera; N State
Organism: Candidatus actinomarina minuta, Homo sapiens, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: GDP, LFA, OLA, RET |
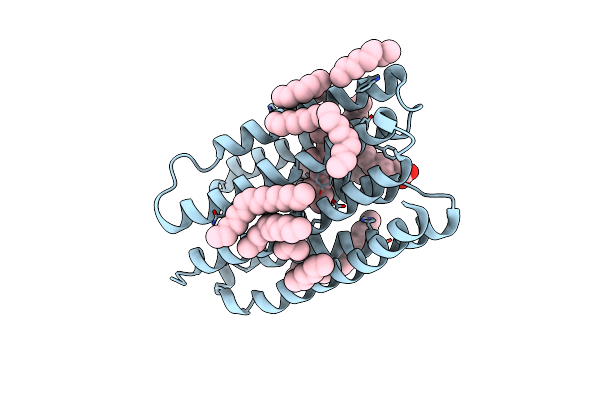 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
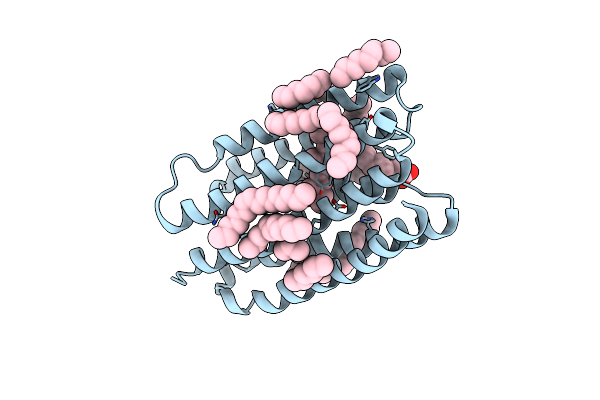
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O* State, Ph 8.8
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
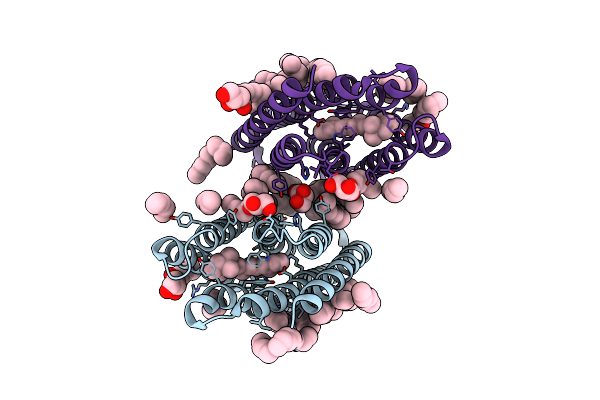
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O State Obtained By Cryotrapping
Organism: Candidatus actinomarina minuta, Marine actinobacteria clade
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: OLA, LFA, RET |
 |
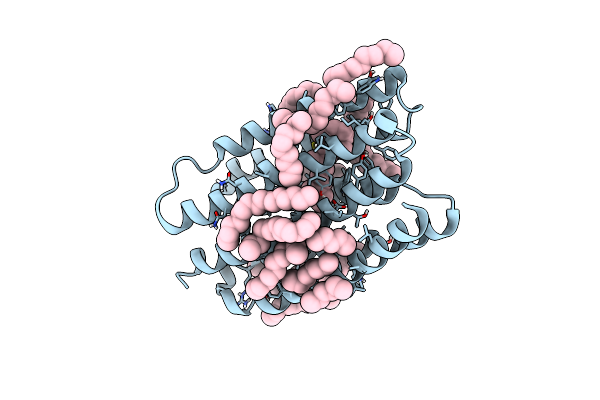
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The M-Like State
Organism: Candidatus actinomarina minuta
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-06-01 Classification: MEMBRANE PROTEIN Ligands: LFA, OLB, OLC, RET |
 |
Crystal Structure Of Marine Actinobacteria Clade Rhodopsin (Mar) In The O State
Organism: Candidatus actinomarina minuta
Method: X-RAY DIFFRACTION Resolution:1.09 Å Release Date: 2022-06-01 Classification: MEMBRANE PROTEIN Ligands: LFA, RET |
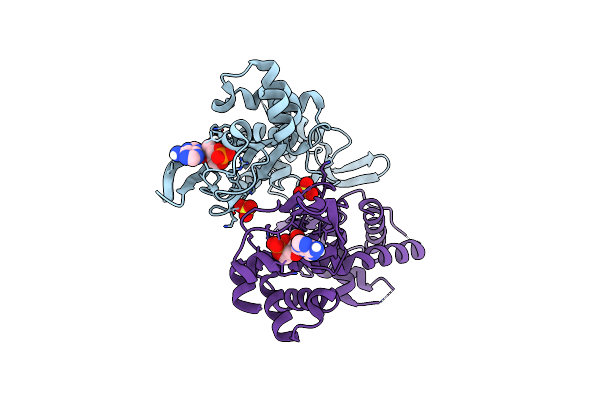 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-02-19 Classification: STRUCTURAL PROTEIN Ligands: CMP, SO4 |
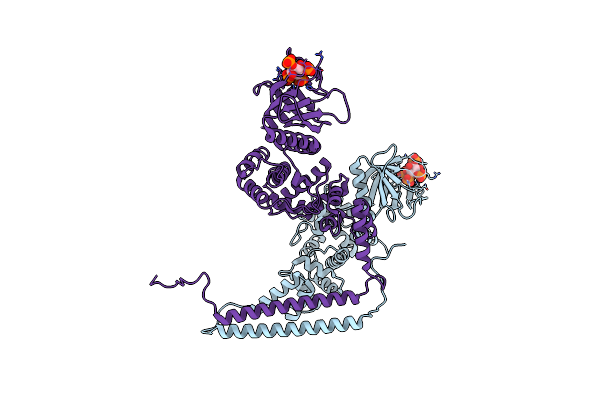 |
Best Fitting Antiparallel Model For Volume 1 Of Truncated Dimeric Cytohesin-3 (Grp1; Amino Acids 14-399)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-09-25 Classification: ENDOCYTOSIS Ligands: 4IP |
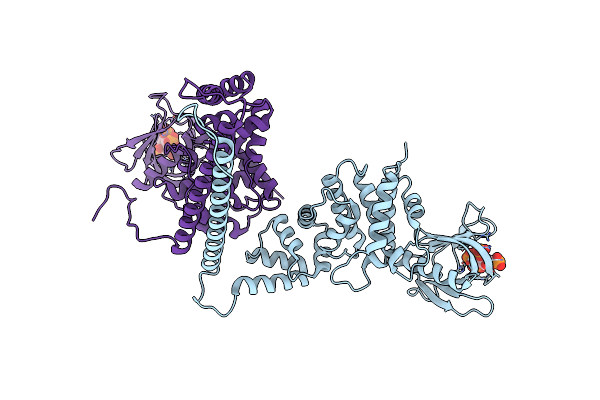 |
Best Fitting Antiparallel Model For Volume 2 Of Truncated Dimeric Cytohesin-3 (Grp1; Amino Acids 14-399)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-09-25 Classification: ENDOCYTOSIS Ligands: 4IP |
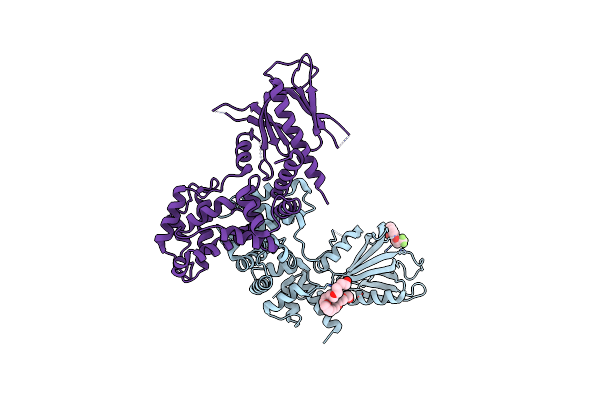 |
Structure Of Human Brag2 (Sec7-Ph Domains) With The Inhibitor Bragsin Bound To The Ph Domain
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-03-13 Classification: HYDROLASE Ligands: DY5, 2PE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2017-12-27 Classification: PROTEIN BINDING Ligands: GDP, MG |
 |
Organism: Legionella pneumophila
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-12-04 Classification: SIGNALING PROTEIN Ligands: GOL |
 |
Organism: Homo sapiens, Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2013-09-25 Classification: PROTEIN TRANSPORT Ligands: G3D |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2012-06-13 Classification: GTP-BINDING PROTEIN Ligands: MG, GDP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2010-08-18 Classification: PROTEIN TRANSPORT Ligands: GDP, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2000-11-13 Classification: OXIDOREDUCTASE Ligands: FAD, SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2000-11-13 Classification: OXIDOREDUCTASE Ligands: FAD, NAP |

