Search Count: 100
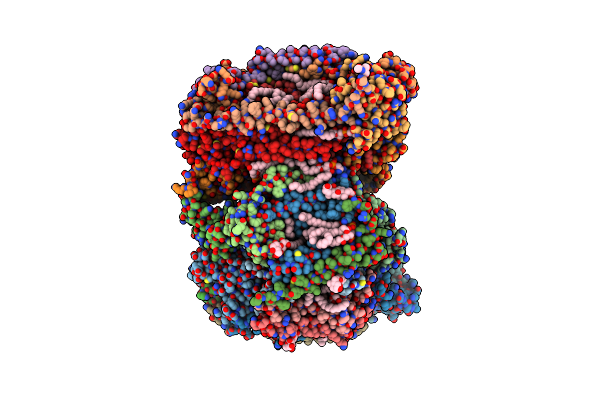 |
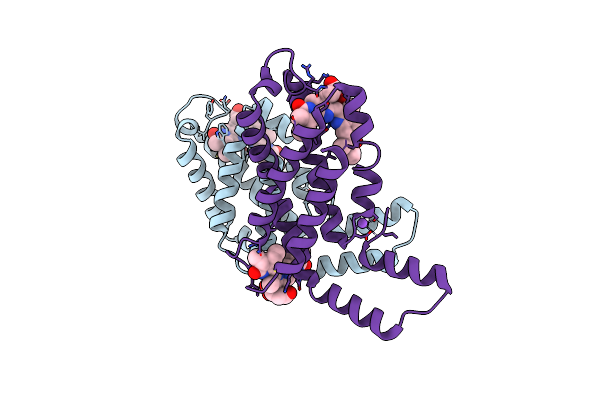
Time-Resolve Sfx Structure Of A Photoproduct Of Carbon Monoxide Complex Of Bovine Cytochrome C Oxidase
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-09-20 Classification: MEMBRANE PROTEIN Ligands: HEA, CU, MG, NA, TGL, PGV, CMO, CUA, CHD, OH, CDL, PSC, ZN, PEK, DMU, SAC |
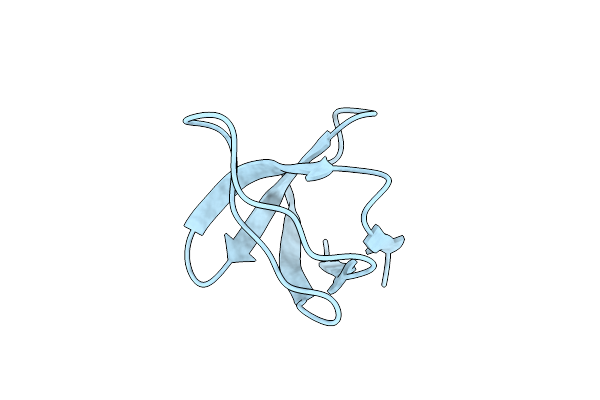 |
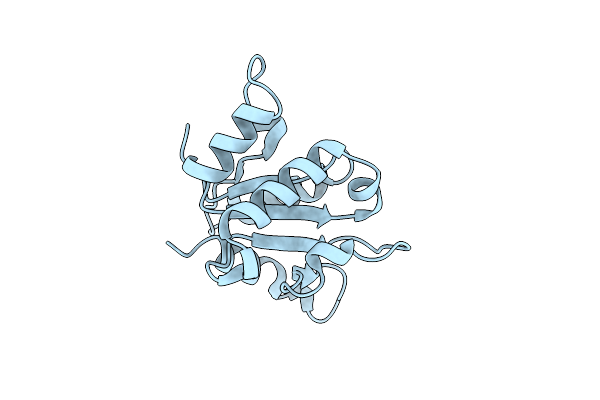
Serial Macromolecular Crystallography At Alba Synchrotron Light Source - Alpha Spectrin-Sh3 Domain
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-10-27 Classification: STRUCTURAL PROTEIN |
 |
Serial Macromolecular Crystallography At Alba Synchrotron Light Source - Lysozyme
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-10-27 Classification: HYDROLASE Ligands: CL |
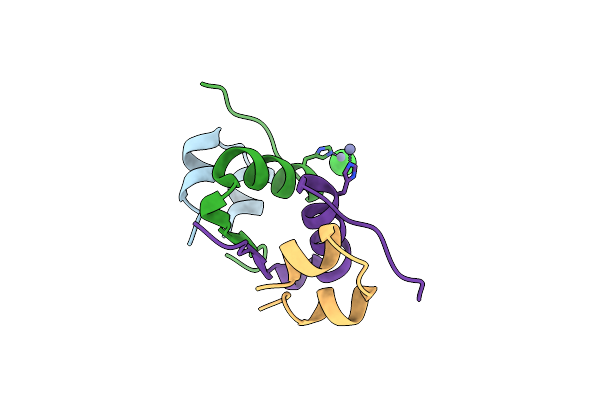 |
Serial Macromolecular Crystallography At Alba Synchrotron Light Source - Insulin
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2021-10-27 Classification: HORMONE Ligands: ZN, CL |
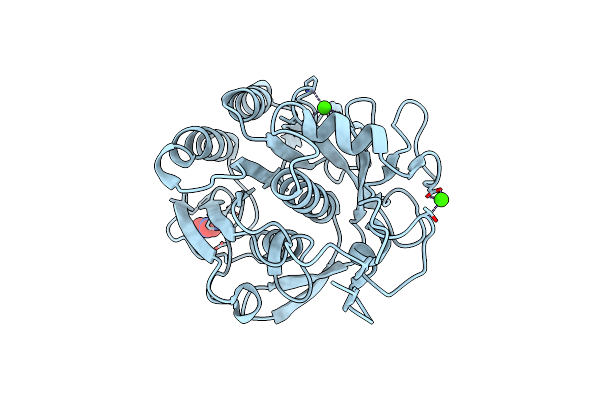 |
Serial Macromolecular Crystallography At Alba Synchrotron Light Source - Proteinase K
Organism: Parengyodontium album
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-10-27 Classification: HYDROLASE Ligands: NO3, CA |
 |
Serial Macromolecular Crystallography At Alba Synchrotron Light Source - Phycocyanin
Organism: Thermosynechococcus vestitus bp-1
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-10-27 Classification: ELECTRON TRANSPORT Ligands: CYC, NA, CL |
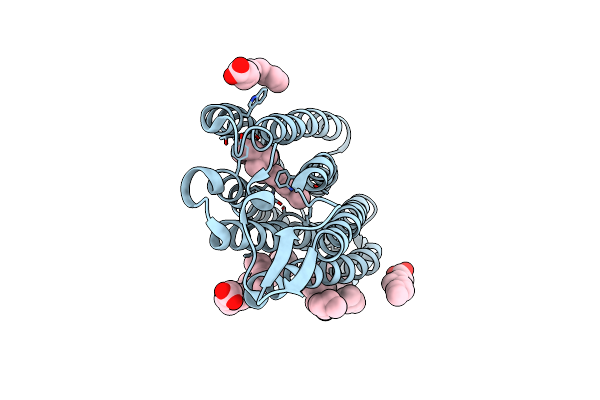 |
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-04-14 Classification: MEMBRANE PROTEIN Ligands: CL, RET, OLA |
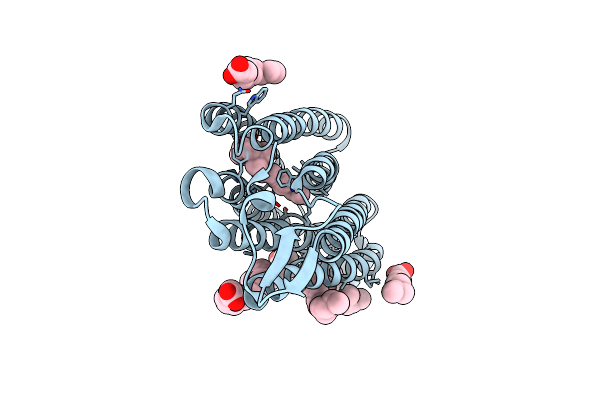 |
Structure Of Chloride Ion Pumping Rhodopsin (Clr) With Ntq Motif 50 Ps After Light Activation
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-04-14 Classification: MEMBRANE PROTEIN Ligands: CL, RET, OLA |
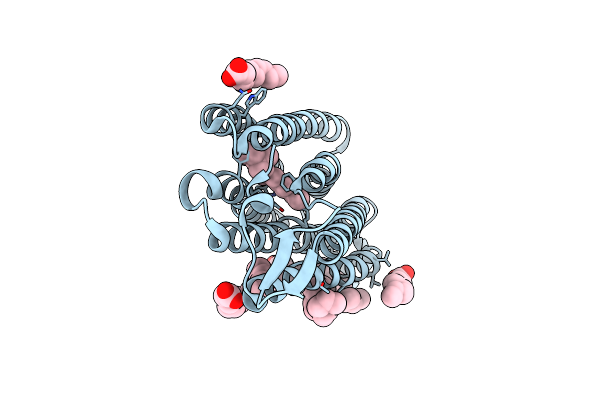 |
Structure Of Chloride Ion Pumping Rhodopsin (Clr) With Ntq Motif 100 Ps After Light Activation (0.90Mj/Mm2)
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-04-14 Classification: MEMBRANE PROTEIN Ligands: CL, RET, OLA |
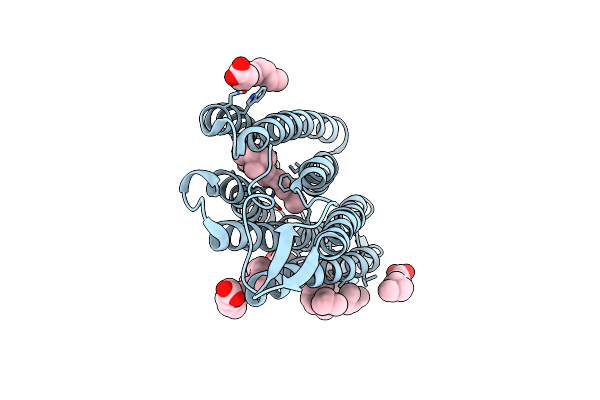 |
Structure Of Chloride Ion Pumping Rhodopsin (Clr) With Ntq Motif 100 Ps After Light Activation (0.17Mj/Mm2)
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-04-14 Classification: MEMBRANE PROTEIN Ligands: CL, RET, OLA |
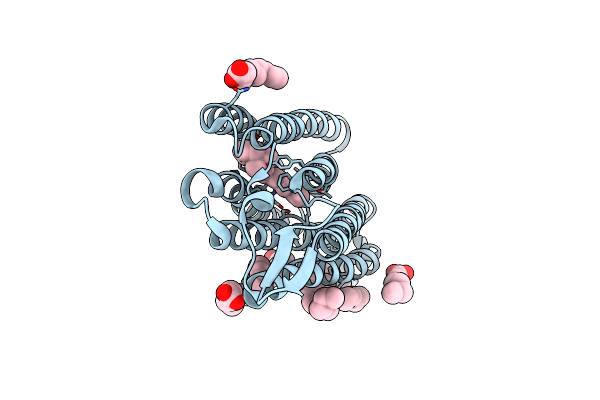 |
Structure Of Chloride Ion Pumping Rhodopsin (Clr) With Ntq Motif 100 Ps After Light Activation (2.63Mj/Mm2)
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-04-14 Classification: MEMBRANE PROTEIN Ligands: CL, RET, OLA |
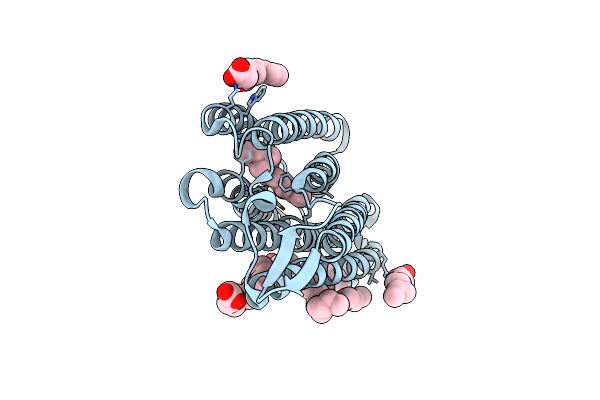 |
Structure Of Chloride Ion Pumping Rhodopsin (Clr) With Ntq Motif 100 Ps After Light Activation (6.49 Mj/Mm2)
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-04-14 Classification: MEMBRANE PROTEIN Ligands: CL, RET, OLA |
 |
Organism: Homo sapiens
Method: ELECTRON CRYSTALLOGRAPHY Resolution:3.00 Å Release Date: 2021-03-10 Classification: PROTEIN BINDING |
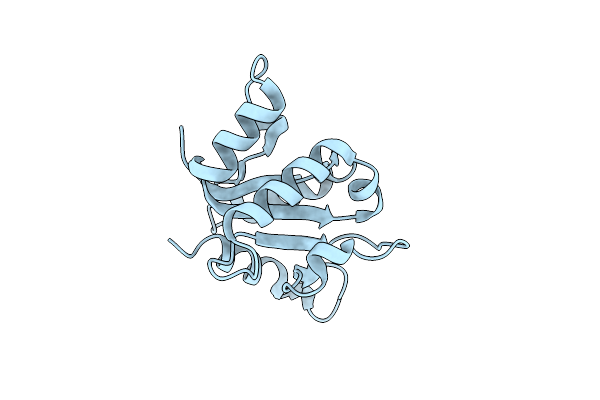 |
Sfx Structure Of The Myd88 Tir Domain Higher-Order Assembly (Solved, Rebuilt And Refined Using An Identical Protocol To The Microed Structure Of The Myd88 Tir Domain Higher-Order Assembly)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-03-10 Classification: PROTEIN BINDING |
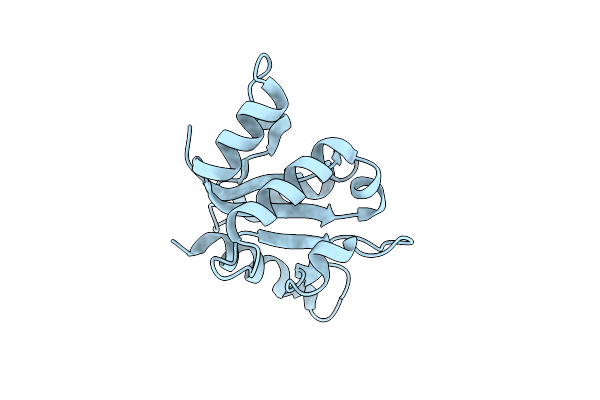 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-03-10 Classification: PROTEIN BINDING |
 |
Transitional Unit Cell 1 Of Adenine Riboswitch Aptamer Crystal Phase Transition Upon Ligand Binding
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2020-11-11 Classification: RNA Ligands: ADE, MG |
 |
Transitional Unit Cell 2 Of Adenine Riboswitch Aptamer Crystal Phase Transition Upon Ligand Binding
Organism: Vibrio vulnificus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2020-11-11 Classification: RNA Ligands: ADE, K, MG |
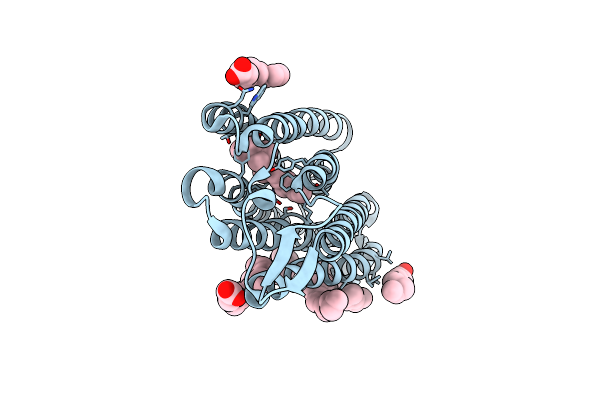 |
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-09-30 Classification: MEMBRANE PROTEIN Ligands: CL, RET, OLA |
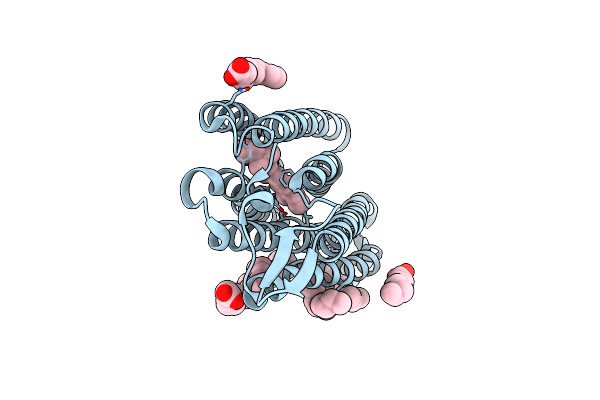 |
Dark State Structure Of Chloride Ion Pumping Rhodopsin (Clr) With Ntq Motif
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-09-16 Classification: MEMBRANE PROTEIN Ligands: CL, RET, OLA |
 |
Organism: Escherichia coli (strain b / bl21-de3)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2020-09-02 Classification: TRANSFERASE |

