Search Count: 21
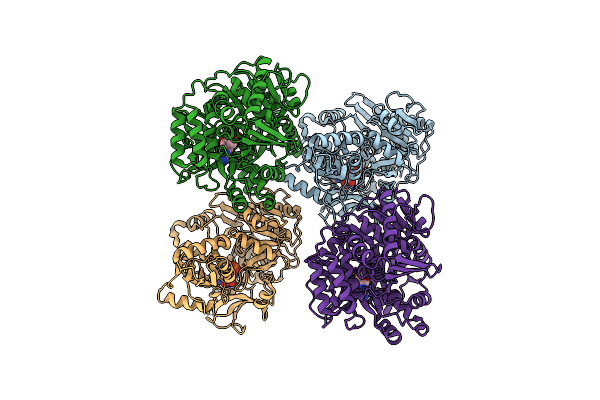 |
Crystal Structure Of Acyl-Coa Synthetase From Metallosphaera Sedula In Complex With Acetyl-Amp
Organism: Metallosphaera sedula dsm 5348
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-11-15 Classification: LIGASE Ligands: AMP, 6R9 |
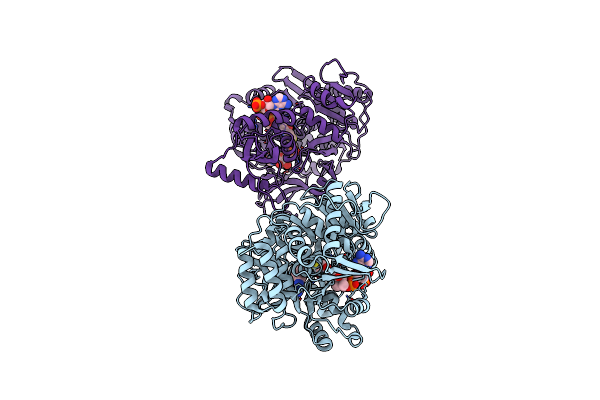 |
Crystal Structure Of Acyl-Coa Synthetase From Metallosphaera Sedula In Complex With Coenzyme A And Acetyl-Amp
Organism: Metallosphaera sedula dsm 5348
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2023-11-15 Classification: LIGASE Ligands: 6R9, COA |
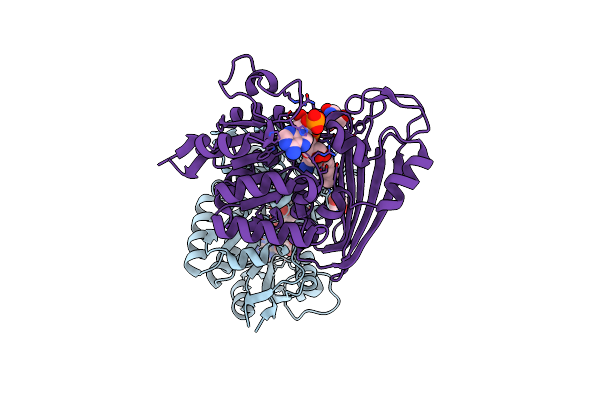 |
Amine Dehydrogenase From Cystobacter Fuscus (Cfusamdh) W145A Mutant With Nadp+ And Pentylamine
Organism: Cystobacter fuscus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: NAP, TRS, AML |
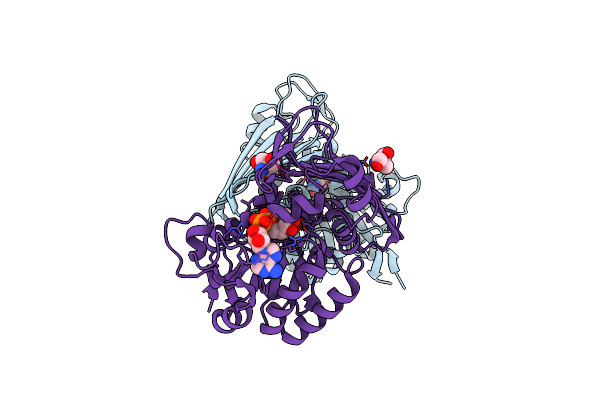 |
Amine Dehydrogenase From Cystobacter Fuscus (Cfusamdh) W145A Mutant With Nad+
Organism: Cystobacter fuscus
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2022-12-07 Classification: OXIDOREDUCTASE Ligands: NAD, TRS |
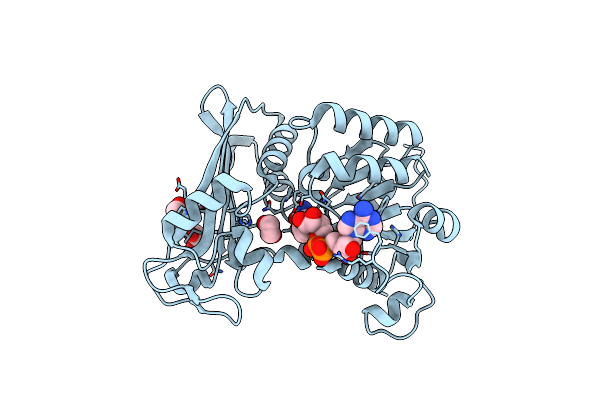 |
Organism: Petrotoga mobilis sj95
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2019-03-27 Classification: OXIDOREDUCTASE Ligands: NAD, EDO |
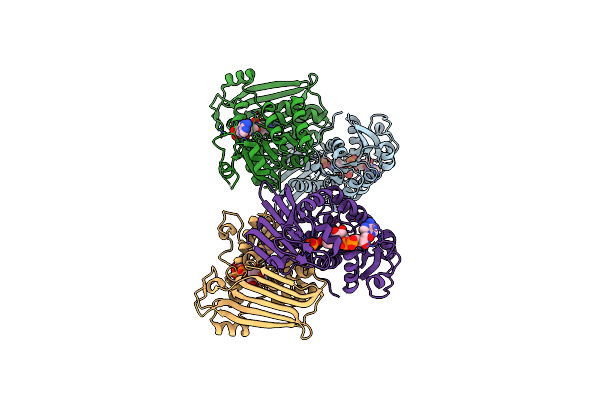 |
Organism: Petrotoga mobilis (strain dsm 10674 / sj95)
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2019-03-27 Classification: OXIDOREDUCTASE Ligands: NAD, PO4 |
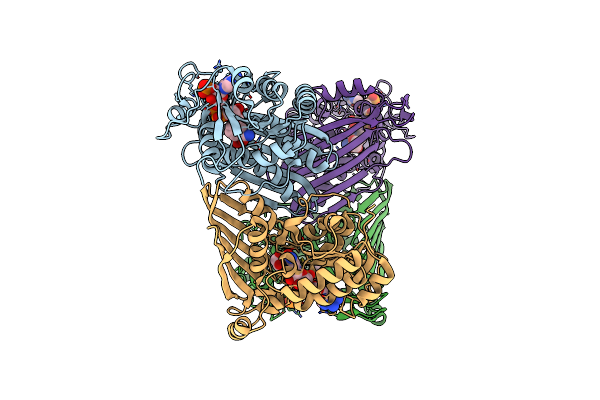 |
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2019-03-27 Classification: OXIDOREDUCTASE Ligands: EDO, NAP |
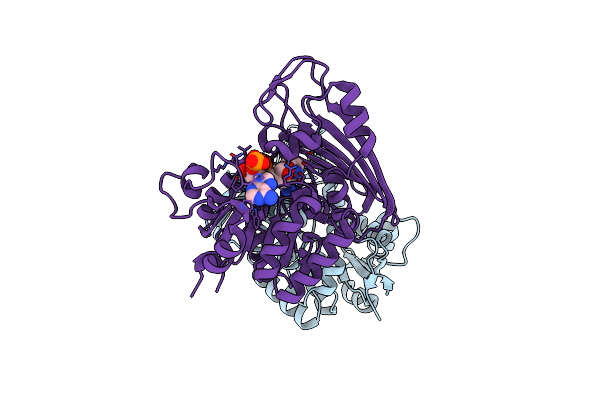 |
Amine Dehydrogenase From Cystobacter Fuscus In Complex With Nadp+ And Cyclohexylamine
Organism: Cystobacter fuscus dsm 2262
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2019-03-27 Classification: OXIDOREDUCTASE Ligands: NAP, HAI |
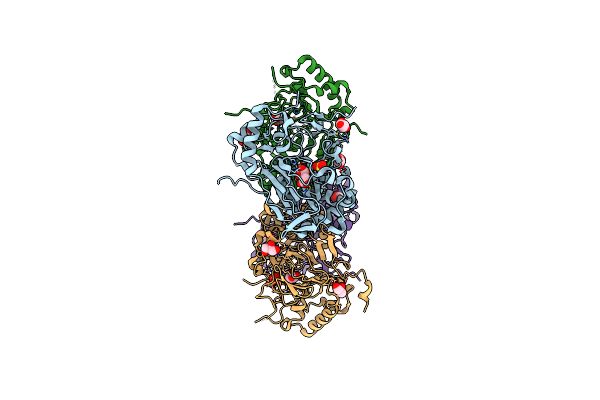 |
Crystal Structure Of Apo Fe(Ii)/Alpha-Ketoglutarate Dependent Dioxygenase Kdo5
Organism: Flavobacterium sp. (strain cf136)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-11-14 Classification: OXIDOREDUCTASE Ligands: FE, MLT, GOL, SO4 |
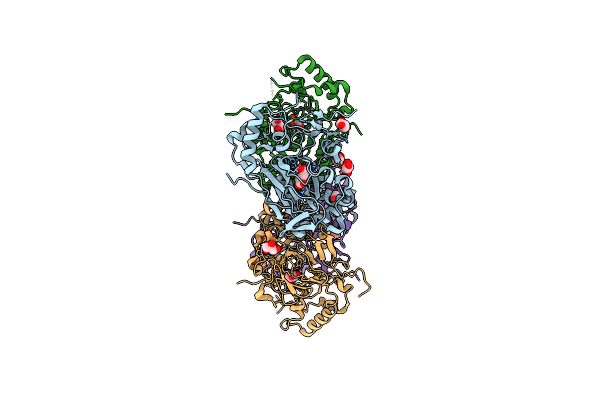 |
Crystal Structure Of The Complex Fe(Ii)/Alpha-Ketoglutarate Dependent Dioxygenase Kdo5 With Fe(Ii)/Alpha-Ketoglutarate
Organism: Flavobacterium sp. (strain cf136)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-11-14 Classification: OXIDOREDUCTASE Ligands: FE, AKG, GOL |
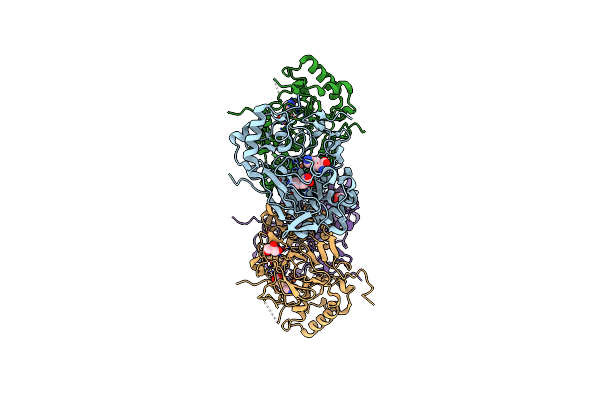 |
Crystal Structure Of The Complex Fe(Ii)/Alpha-Ketoglutarate Dependent Dioxygenase Kdo5 With Fe(Ii)/Lysine
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-11-14 Classification: OXIDOREDUCTASE Ligands: FE, LYS, GOL |
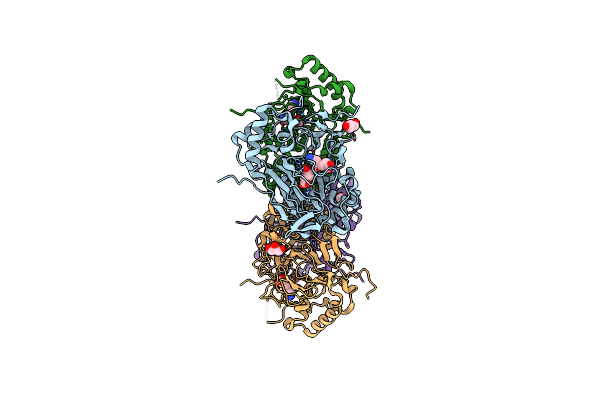 |
Crystal Structure Of The Complex Fe(Ii)/Alpha-Ketoglutarate Dependent Dioxygenase Kdo5 With Fe(Ii)/Succinate/(4R)-4-Hydroxy-L-Lysine
Organism: Flavobacterium sp.
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-11-14 Classification: OXIDOREDUCTASE Ligands: FE, LYO, SIN, GOL |
 |
Crystal Structure Of The Complex Fe(Ii)/Alpha-Ketoglutarate Dependent Dioxygenase Kdo1 With Fe(Ii)/Lysine
Organism: Catenulispora acidiphila (strain dsm 44928 / nrrl b-24433 / nbrc 102108 / jcm 14897)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-11-14 Classification: OXIDOREDUCTASE Ligands: FE2, LYS, ACY, CL |
 |
Crystal Structure Of The Complex Fe(Ii)/Alpha-Ketoglutarate Dependent Dioxygenase Kdo1 With Fe(Ii)/Alpha-Ketoglutarate
Organism: Catenulispora acidiphila (strain dsm 44928 / nrrl b-24433 / nbrc 102108 / jcm 14897)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-11-14 Classification: OXIDOREDUCTASE Ligands: FE2, AKG |
 |
Crystal Structure Of The Apo Fe(Ii)/Alpha-Ketoglutarate Dependent Dioxygenase Kdo1
Organism: Catenulispora acidiphila (strain dsm 44928 / nrrl b-24433 / nbrc 102108 / jcm 14897)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-11-14 Classification: OXIDOREDUCTASE Ligands: ACY |
 |
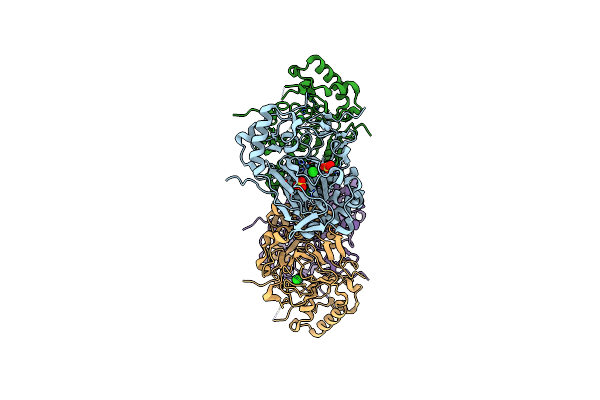
Crystal Structure Of The Fe(Ii)/Alpha-Ketoglutarate Dependent Dioxygenase Kdo1 With Fe(Ii)/Succinate/(3S)-3-Hydroxy-L-Lysine
Organism: Catenulispora acidiphila (strain dsm 44928 / nrrl b-24433 / nbrc 102108 / jcm 14897)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-11-14 Classification: OXIDOREDUCTASE Ligands: FE, SIN, CUW, ACT |
 |
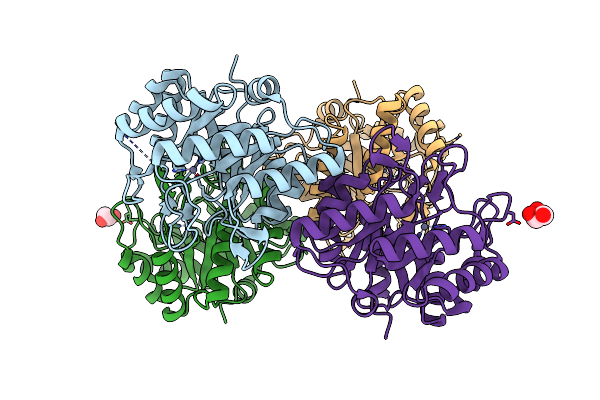
Crystal Structure Of The Fe(Ii)/Alpha-Ketoglutarate Dependent Dioxygenase Kdo5 With Re(Ii)
Organism: Flavobacterium sp. (strain cf136)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-11-14 Classification: OXIDOREDUCTASE Ligands: CL, RE, SO4, GOL |
 |
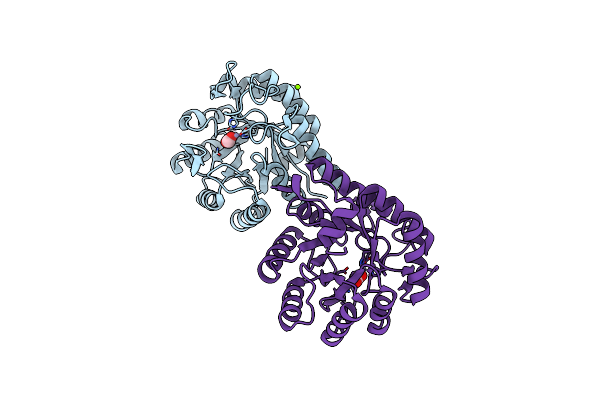
Crystal Structure Of The 3-Keto-5-Aminohexanoate Cleavage Enzyme (Kce) From Candidatus Cloacamonas Acidaminovorans (Orthorombic Form)
Organism: Candidatus cloacamonas acidaminovorans
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2011-06-01 Classification: LYASE Ligands: ZN, GOL |
 |
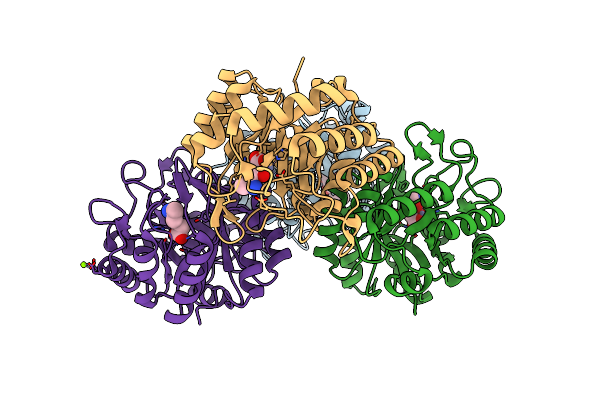
Crystal Structure Of The 3-Keto-5-Aminohexanoate Cleavage Enzyme (Kce) From Candidatus Cloacamonas Acidaminovorans (Tetragonal Form)
Organism: Candidatus cloacamonas acidaminovorans
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2011-06-01 Classification: LYASE Ligands: ZN, FMT, MG |
 |
Crystal Structure Of The 3-Keto-5-Aminohexanoate Cleavage Enzyme (Kce) From C. Cloacamonas Acidaminovorans In Complex With The Substrate 3- Keto-5-Aminohexanoate
Organism: Candidatus cloacamonas acidaminovorans
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2011-06-01 Classification: LYASE Ligands: ZN, KMH, MG |

